
Cereal yellow dwarf virus RPV
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Polerovirus
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins
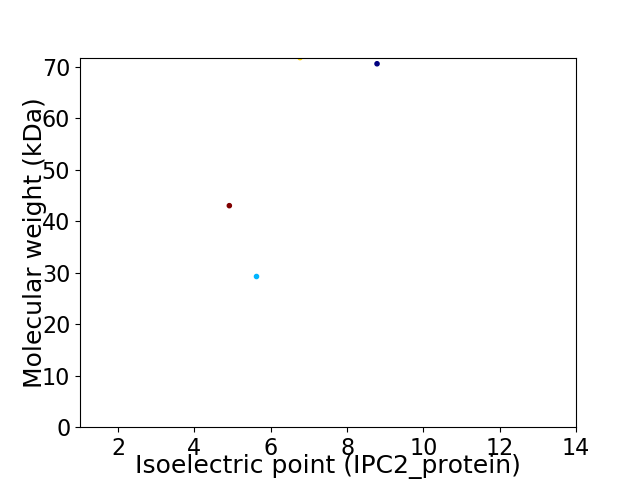
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q65863|Q65863_9LUTE RNA-directed RNA polymerase (Fragment) OS=Cereal yellow dwarf virus RPV OX=2170100 PE=4 SV=1
VV1 pKa = 7.21DD2 pKa = 4.96AEE4 pKa = 4.77PGPSPGPSPDD14 pKa = 3.76PPPPPSPSPEE24 pKa = 3.7PAPAKK29 pKa = 9.1EE30 pKa = 3.96EE31 pKa = 4.0RR32 pKa = 11.84FIVYY36 pKa = 9.99SGVAHH41 pKa = 6.68TIISAQSTDD50 pKa = 2.88DD51 pKa = 3.97SIIVRR56 pKa = 11.84DD57 pKa = 3.68IPDD60 pKa = 2.93QRR62 pKa = 11.84FRR64 pKa = 11.84YY65 pKa = 9.42VEE67 pKa = 3.84NEE69 pKa = 3.31NFYY72 pKa = 10.01WFQIAAQWYY81 pKa = 9.07SNTNTKK87 pKa = 9.69AVPMFVFPVPIGEE100 pKa = 4.08WSVEE104 pKa = 3.66ISTEE108 pKa = 4.42GYY110 pKa = 9.58QATSSTTDD118 pKa = 3.54PNKK121 pKa = 10.51GRR123 pKa = 11.84IDD125 pKa = 3.54GLIAYY130 pKa = 7.63DD131 pKa = 3.86NSSEE135 pKa = 3.83GWNIGAGSNVTITNNKK151 pKa = 9.13ADD153 pKa = 3.8NSWKK157 pKa = 10.41YY158 pKa = 9.45GHH160 pKa = 7.74PDD162 pKa = 3.24LEE164 pKa = 4.45INSCHH169 pKa = 6.41FNQNQVLEE177 pKa = 4.13KK178 pKa = 10.75DD179 pKa = 4.3GIISFHH185 pKa = 6.15VKK187 pKa = 9.04ATEE190 pKa = 3.83KK191 pKa = 10.18EE192 pKa = 4.23ANFFLVAPPVQKK204 pKa = 9.64TSKK207 pKa = 9.54YY208 pKa = 9.66NYY210 pKa = 8.82AVSYY214 pKa = 8.92GAWTDD219 pKa = 2.96RR220 pKa = 11.84DD221 pKa = 3.91MEE223 pKa = 4.8FGLITVTLDD232 pKa = 3.13EE233 pKa = 4.77KK234 pKa = 10.74RR235 pKa = 11.84GSGSPTRR242 pKa = 11.84KK243 pKa = 9.56SLRR246 pKa = 11.84AGHH249 pKa = 6.75AGVTTTTDD257 pKa = 3.42LVALPEE263 pKa = 4.23MEE265 pKa = 3.91NSGIEE270 pKa = 4.11TSEE273 pKa = 4.26TPSAPVTSSKK283 pKa = 11.25APLPTVSDD291 pKa = 4.22SEE293 pKa = 5.19SEE295 pKa = 4.19DD296 pKa = 4.15DD297 pKa = 4.59PLSAAPDD304 pKa = 3.45VGFGGTRR311 pKa = 11.84LLIDD315 pKa = 3.93TDD317 pKa = 3.28IKK319 pKa = 10.14TIPDD323 pKa = 4.07PDD325 pKa = 3.62VADD328 pKa = 3.96AFVNSAHH335 pKa = 6.54VGVDD339 pKa = 2.97PWAEE343 pKa = 3.6VRR345 pKa = 11.84AFKK348 pKa = 10.48RR349 pKa = 11.84AQRR352 pKa = 11.84PPRR355 pKa = 11.84GPSSVASSSLSGGSLRR371 pKa = 11.84GSLRR375 pKa = 11.84PKK377 pKa = 10.28TEE379 pKa = 3.9DD380 pKa = 3.55PKK382 pKa = 11.36DD383 pKa = 3.65SSKK386 pKa = 11.25SKK388 pKa = 9.64SRR390 pKa = 11.84KK391 pKa = 7.92WSLGSLRR398 pKa = 4.35
VV1 pKa = 7.21DD2 pKa = 4.96AEE4 pKa = 4.77PGPSPGPSPDD14 pKa = 3.76PPPPPSPSPEE24 pKa = 3.7PAPAKK29 pKa = 9.1EE30 pKa = 3.96EE31 pKa = 4.0RR32 pKa = 11.84FIVYY36 pKa = 9.99SGVAHH41 pKa = 6.68TIISAQSTDD50 pKa = 2.88DD51 pKa = 3.97SIIVRR56 pKa = 11.84DD57 pKa = 3.68IPDD60 pKa = 2.93QRR62 pKa = 11.84FRR64 pKa = 11.84YY65 pKa = 9.42VEE67 pKa = 3.84NEE69 pKa = 3.31NFYY72 pKa = 10.01WFQIAAQWYY81 pKa = 9.07SNTNTKK87 pKa = 9.69AVPMFVFPVPIGEE100 pKa = 4.08WSVEE104 pKa = 3.66ISTEE108 pKa = 4.42GYY110 pKa = 9.58QATSSTTDD118 pKa = 3.54PNKK121 pKa = 10.51GRR123 pKa = 11.84IDD125 pKa = 3.54GLIAYY130 pKa = 7.63DD131 pKa = 3.86NSSEE135 pKa = 3.83GWNIGAGSNVTITNNKK151 pKa = 9.13ADD153 pKa = 3.8NSWKK157 pKa = 10.41YY158 pKa = 9.45GHH160 pKa = 7.74PDD162 pKa = 3.24LEE164 pKa = 4.45INSCHH169 pKa = 6.41FNQNQVLEE177 pKa = 4.13KK178 pKa = 10.75DD179 pKa = 4.3GIISFHH185 pKa = 6.15VKK187 pKa = 9.04ATEE190 pKa = 3.83KK191 pKa = 10.18EE192 pKa = 4.23ANFFLVAPPVQKK204 pKa = 9.64TSKK207 pKa = 9.54YY208 pKa = 9.66NYY210 pKa = 8.82AVSYY214 pKa = 8.92GAWTDD219 pKa = 2.96RR220 pKa = 11.84DD221 pKa = 3.91MEE223 pKa = 4.8FGLITVTLDD232 pKa = 3.13EE233 pKa = 4.77KK234 pKa = 10.74RR235 pKa = 11.84GSGSPTRR242 pKa = 11.84KK243 pKa = 9.56SLRR246 pKa = 11.84AGHH249 pKa = 6.75AGVTTTTDD257 pKa = 3.42LVALPEE263 pKa = 4.23MEE265 pKa = 3.91NSGIEE270 pKa = 4.11TSEE273 pKa = 4.26TPSAPVTSSKK283 pKa = 11.25APLPTVSDD291 pKa = 4.22SEE293 pKa = 5.19SEE295 pKa = 4.19DD296 pKa = 4.15DD297 pKa = 4.59PLSAAPDD304 pKa = 3.45VGFGGTRR311 pKa = 11.84LLIDD315 pKa = 3.93TDD317 pKa = 3.28IKK319 pKa = 10.14TIPDD323 pKa = 4.07PDD325 pKa = 3.62VADD328 pKa = 3.96AFVNSAHH335 pKa = 6.54VGVDD339 pKa = 2.97PWAEE343 pKa = 3.6VRR345 pKa = 11.84AFKK348 pKa = 10.48RR349 pKa = 11.84AQRR352 pKa = 11.84PPRR355 pKa = 11.84GPSSVASSSLSGGSLRR371 pKa = 11.84GSLRR375 pKa = 11.84PKK377 pKa = 10.28TEE379 pKa = 3.9DD380 pKa = 3.55PKK382 pKa = 11.36DD383 pKa = 3.65SSKK386 pKa = 11.25SKK388 pKa = 9.64SRR390 pKa = 11.84KK391 pKa = 7.92WSLGSLRR398 pKa = 4.35
Molecular weight: 43.01 kDa
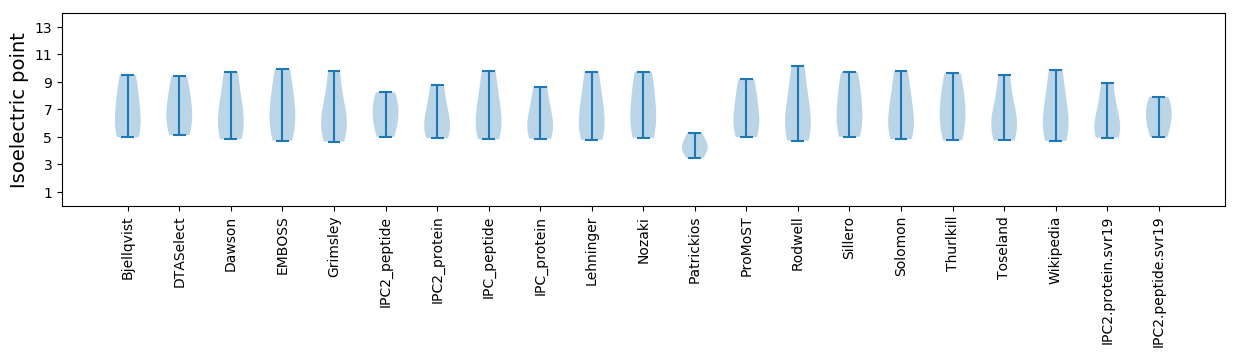
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q89645|Q89645_9LUTE Serine protease OS=Cereal yellow dwarf virus RPV OX=2170100 PE=4 SV=1
MM1 pKa = 7.25KK2 pKa = 10.27SIYY5 pKa = 9.6FVCLLAFCCQFSSQEE20 pKa = 3.91NLTLGAVTLTSPFLTPNYY38 pKa = 9.88LFDD41 pKa = 5.76GGSQWGTSLPSPLMVTFDD59 pKa = 3.49RR60 pKa = 11.84TQNSASTCPPCQPALSTSSSYY81 pKa = 11.7SDD83 pKa = 3.95IIQVGWQKK91 pKa = 11.26LSLDD95 pKa = 3.73SKK97 pKa = 10.77SAFKK101 pKa = 10.67AAWSLLKK108 pKa = 10.62DD109 pKa = 3.41SCTTASRR116 pKa = 11.84ALKK119 pKa = 10.36AAFHH123 pKa = 6.73DD124 pKa = 4.55LVGKK128 pKa = 9.81ALWLLVLIWTGLLRR142 pKa = 11.84QLFSAVWSAITNYY155 pKa = 10.47SLPVCLLISLGIITSWLWKK174 pKa = 8.42ACRR177 pKa = 11.84WLFGTLPALLCITLVKK193 pKa = 10.72NIFRR197 pKa = 11.84ILTFKK202 pKa = 10.82RR203 pKa = 11.84FFNEE207 pKa = 4.1KK208 pKa = 8.7TVSGYY213 pKa = 10.89DD214 pKa = 3.36SYY216 pKa = 11.55SIPSTPPKK224 pKa = 10.01RR225 pKa = 11.84SVIMMRR231 pKa = 11.84RR232 pKa = 11.84QNKK235 pKa = 7.03EE236 pKa = 3.55HH237 pKa = 6.97IGYY240 pKa = 9.95ANCIRR245 pKa = 11.84LFDD248 pKa = 3.92GRR250 pKa = 11.84NAIVTVAHH258 pKa = 6.67NIEE261 pKa = 4.54EE262 pKa = 4.66GCSFYY267 pKa = 11.16SSRR270 pKa = 11.84TSGSIPITEE279 pKa = 3.72FRR281 pKa = 11.84VIFEE285 pKa = 4.51SKK287 pKa = 9.85TMDD290 pKa = 3.0IAILVGPINWEE301 pKa = 4.18SILGCKK307 pKa = 9.29GVHH310 pKa = 5.5FTTADD315 pKa = 3.59RR316 pKa = 11.84LAEE319 pKa = 4.4CPAALYY325 pKa = 11.03LLDD328 pKa = 5.86SDD330 pKa = 4.52GQWRR334 pKa = 11.84SNSAKK339 pKa = 10.13ICGHH343 pKa = 6.57FDD345 pKa = 2.86NFAQVLSNTKK355 pKa = 9.69VGHH358 pKa = 5.97SGAGYY363 pKa = 9.82FYY365 pKa = 11.05GKK367 pKa = 8.15TLVGLHH373 pKa = 6.6KK374 pKa = 10.39GHH376 pKa = 7.46PGKK379 pKa = 10.42DD380 pKa = 3.41FNFNLMAPLPGIPGLTSPQYY400 pKa = 10.78VVEE403 pKa = 4.26SDD405 pKa = 3.71PPQGLVFPEE414 pKa = 4.23EE415 pKa = 4.04VTEE418 pKa = 4.45SIEE421 pKa = 4.19AAIKK425 pKa = 9.36EE426 pKa = 3.77ATMYY430 pKa = 11.23KK431 pKa = 10.38NVFANRR437 pKa = 11.84GRR439 pKa = 11.84GAFKK443 pKa = 10.64SKK445 pKa = 10.81SGINWEE451 pKa = 4.64DD452 pKa = 3.62IEE454 pKa = 5.35DD455 pKa = 3.56EE456 pKa = 4.33SGNGKK461 pKa = 9.44AAASAVTNAAAANKK475 pKa = 9.58VIATPGVAKK484 pKa = 10.09SQKK487 pKa = 8.23KK488 pKa = 6.64TAVPSSPKK496 pKa = 10.39APQPPAASQPTSTTGSRR513 pKa = 11.84TPICPIATATSPDD526 pKa = 3.52TAVPTGPSQQEE537 pKa = 3.57IMNNIMNLLVQRR549 pKa = 11.84IDD551 pKa = 3.11MSKK554 pKa = 9.37IEE556 pKa = 4.35KK557 pKa = 10.58SIVDD561 pKa = 3.38QVANQALKK569 pKa = 10.45KK570 pKa = 9.95PRR572 pKa = 11.84GKK574 pKa = 10.25RR575 pKa = 11.84GSKK578 pKa = 9.59KK579 pKa = 10.01RR580 pKa = 11.84PAAGKK585 pKa = 9.92SSSPTSTPGTYY596 pKa = 8.5QHH598 pKa = 7.01PNKK601 pKa = 10.03KK602 pKa = 9.41SQASNRR608 pKa = 11.84LGNSPPSTTRR618 pKa = 11.84APAANQNGGEE628 pKa = 3.9NSVPNTLSWVRR639 pKa = 11.84KK640 pKa = 9.05LPVSGGPKK648 pKa = 9.69SGHH651 pKa = 6.36PLNN654 pKa = 4.74
MM1 pKa = 7.25KK2 pKa = 10.27SIYY5 pKa = 9.6FVCLLAFCCQFSSQEE20 pKa = 3.91NLTLGAVTLTSPFLTPNYY38 pKa = 9.88LFDD41 pKa = 5.76GGSQWGTSLPSPLMVTFDD59 pKa = 3.49RR60 pKa = 11.84TQNSASTCPPCQPALSTSSSYY81 pKa = 11.7SDD83 pKa = 3.95IIQVGWQKK91 pKa = 11.26LSLDD95 pKa = 3.73SKK97 pKa = 10.77SAFKK101 pKa = 10.67AAWSLLKK108 pKa = 10.62DD109 pKa = 3.41SCTTASRR116 pKa = 11.84ALKK119 pKa = 10.36AAFHH123 pKa = 6.73DD124 pKa = 4.55LVGKK128 pKa = 9.81ALWLLVLIWTGLLRR142 pKa = 11.84QLFSAVWSAITNYY155 pKa = 10.47SLPVCLLISLGIITSWLWKK174 pKa = 8.42ACRR177 pKa = 11.84WLFGTLPALLCITLVKK193 pKa = 10.72NIFRR197 pKa = 11.84ILTFKK202 pKa = 10.82RR203 pKa = 11.84FFNEE207 pKa = 4.1KK208 pKa = 8.7TVSGYY213 pKa = 10.89DD214 pKa = 3.36SYY216 pKa = 11.55SIPSTPPKK224 pKa = 10.01RR225 pKa = 11.84SVIMMRR231 pKa = 11.84RR232 pKa = 11.84QNKK235 pKa = 7.03EE236 pKa = 3.55HH237 pKa = 6.97IGYY240 pKa = 9.95ANCIRR245 pKa = 11.84LFDD248 pKa = 3.92GRR250 pKa = 11.84NAIVTVAHH258 pKa = 6.67NIEE261 pKa = 4.54EE262 pKa = 4.66GCSFYY267 pKa = 11.16SSRR270 pKa = 11.84TSGSIPITEE279 pKa = 3.72FRR281 pKa = 11.84VIFEE285 pKa = 4.51SKK287 pKa = 9.85TMDD290 pKa = 3.0IAILVGPINWEE301 pKa = 4.18SILGCKK307 pKa = 9.29GVHH310 pKa = 5.5FTTADD315 pKa = 3.59RR316 pKa = 11.84LAEE319 pKa = 4.4CPAALYY325 pKa = 11.03LLDD328 pKa = 5.86SDD330 pKa = 4.52GQWRR334 pKa = 11.84SNSAKK339 pKa = 10.13ICGHH343 pKa = 6.57FDD345 pKa = 2.86NFAQVLSNTKK355 pKa = 9.69VGHH358 pKa = 5.97SGAGYY363 pKa = 9.82FYY365 pKa = 11.05GKK367 pKa = 8.15TLVGLHH373 pKa = 6.6KK374 pKa = 10.39GHH376 pKa = 7.46PGKK379 pKa = 10.42DD380 pKa = 3.41FNFNLMAPLPGIPGLTSPQYY400 pKa = 10.78VVEE403 pKa = 4.26SDD405 pKa = 3.71PPQGLVFPEE414 pKa = 4.23EE415 pKa = 4.04VTEE418 pKa = 4.45SIEE421 pKa = 4.19AAIKK425 pKa = 9.36EE426 pKa = 3.77ATMYY430 pKa = 11.23KK431 pKa = 10.38NVFANRR437 pKa = 11.84GRR439 pKa = 11.84GAFKK443 pKa = 10.64SKK445 pKa = 10.81SGINWEE451 pKa = 4.64DD452 pKa = 3.62IEE454 pKa = 5.35DD455 pKa = 3.56EE456 pKa = 4.33SGNGKK461 pKa = 9.44AAASAVTNAAAANKK475 pKa = 9.58VIATPGVAKK484 pKa = 10.09SQKK487 pKa = 8.23KK488 pKa = 6.64TAVPSSPKK496 pKa = 10.39APQPPAASQPTSTTGSRR513 pKa = 11.84TPICPIATATSPDD526 pKa = 3.52TAVPTGPSQQEE537 pKa = 3.57IMNNIMNLLVQRR549 pKa = 11.84IDD551 pKa = 3.11MSKK554 pKa = 9.37IEE556 pKa = 4.35KK557 pKa = 10.58SIVDD561 pKa = 3.38QVANQALKK569 pKa = 10.45KK570 pKa = 9.95PRR572 pKa = 11.84GKK574 pKa = 10.25RR575 pKa = 11.84GSKK578 pKa = 9.59KK579 pKa = 10.01RR580 pKa = 11.84PAAGKK585 pKa = 9.92SSSPTSTPGTYY596 pKa = 8.5QHH598 pKa = 7.01PNKK601 pKa = 10.03KK602 pKa = 9.41SQASNRR608 pKa = 11.84LGNSPPSTTRR618 pKa = 11.84APAANQNGGEE628 pKa = 3.9NSVPNTLSWVRR639 pKa = 11.84KK640 pKa = 9.05LPVSGGPKK648 pKa = 9.69SGHH651 pKa = 6.36PLNN654 pKa = 4.74
Molecular weight: 70.57 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1941 |
256 |
654 |
485.3 |
53.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.676 ± 0.509 | 2.009 ± 0.543 |
4.688 ± 0.8 | 5.564 ± 0.956 |
4.122 ± 0.651 | 6.801 ± 0.206 |
1.958 ± 0.268 | 4.688 ± 0.565 |
4.946 ± 0.773 | 8.501 ± 1.368 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.443 ± 0.209 | 4.482 ± 0.366 |
6.698 ± 1.002 | 3.658 ± 0.449 |
5.77 ± 1.034 | 9.995 ± 0.895 |
6.182 ± 0.6 | 5.925 ± 0.295 |
2.164 ± 0.186 | 2.731 ± 0.425 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
