
Trichoderma harzianum bipartite mycovirus 1
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Curvulaviridae; Orthocurvulavirus; Trichoderma harzianum orthocurvulavirus 1
Average proteome isoelectric point is 7.27
Get precalculated fractions of proteins
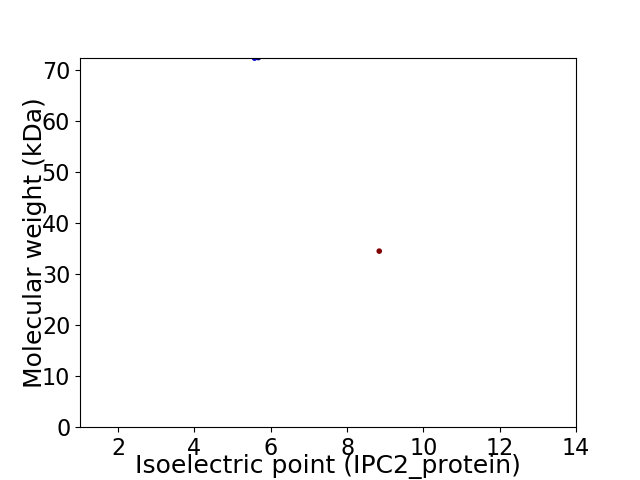
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
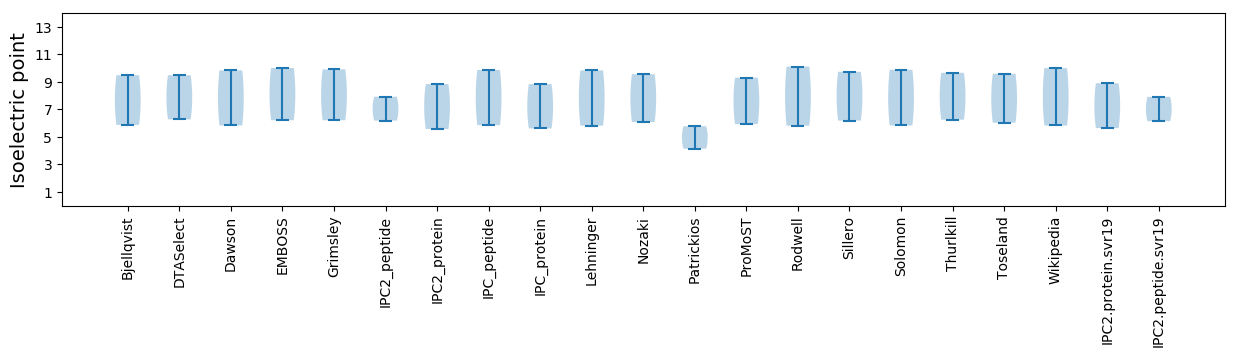
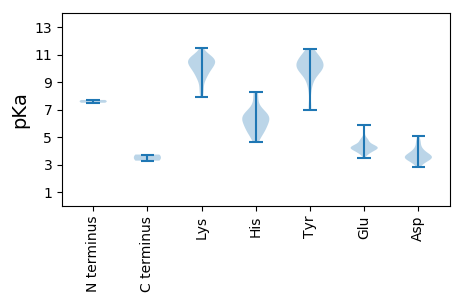
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A346THI6|A0A346THI6_9VIRU Uncharacterized protein OS=Trichoderma harzianum bipartite mycovirus 1 OX=2315392 PE=4 SV=1
MM1 pKa = 7.49NAPEE5 pKa = 4.12IPGRR9 pKa = 11.84NPDD12 pKa = 3.83RR13 pKa = 11.84IRR15 pKa = 11.84RR16 pKa = 11.84SSLFNFDD23 pKa = 4.26GGLLTDD29 pKa = 3.2SHH31 pKa = 8.25GDD33 pKa = 3.31VLLNTRR39 pKa = 11.84KK40 pKa = 10.04RR41 pKa = 11.84GRR43 pKa = 11.84NQFDD47 pKa = 4.42DD48 pKa = 3.93EE49 pKa = 4.88TEE51 pKa = 4.26VTSAPSSHH59 pKa = 6.43TSGNASTRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 9.35RR71 pKa = 11.84LRR73 pKa = 11.84RR74 pKa = 11.84VRR76 pKa = 11.84MASHH80 pKa = 6.35HH81 pKa = 5.6TDD83 pKa = 2.95EE84 pKa = 4.8MSALTKK90 pKa = 10.41LFQHH94 pKa = 6.66ISILGRR100 pKa = 11.84IEE102 pKa = 4.09QYY104 pKa = 11.38NKK106 pKa = 10.49LNPDD110 pKa = 3.97LVVADD115 pKa = 4.66PDD117 pKa = 4.13PLIVEE122 pKa = 4.56YY123 pKa = 9.84TLHH126 pKa = 6.48NPPIIPNFKK135 pKa = 10.02PEE137 pKa = 4.74DD138 pKa = 3.62YY139 pKa = 10.18TFVKK143 pKa = 10.58ACTPVEE149 pKa = 4.1MAHH152 pKa = 6.73LKK154 pKa = 10.74HH155 pKa = 6.62FDD157 pKa = 3.59RR158 pKa = 11.84EE159 pKa = 4.18EE160 pKa = 4.14SVPQAEE166 pKa = 4.75YY167 pKa = 10.9LPSLQTASDD176 pKa = 3.46IVGKK180 pKa = 10.17LLSLPEE186 pKa = 3.94YY187 pKa = 10.87LFFPEE192 pKa = 5.42VDD194 pKa = 3.45DD195 pKa = 4.73LNNVRR200 pKa = 11.84YY201 pKa = 9.68IGSKK205 pKa = 10.41FPGIEE210 pKa = 3.76YY211 pKa = 10.72AMQGFKK217 pKa = 9.43TRR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.04AHH223 pKa = 5.73EE224 pKa = 3.99MAMTDD229 pKa = 3.52ALSAFDD235 pKa = 4.29KK236 pKa = 10.76LLNGEE241 pKa = 4.19RR242 pKa = 11.84VKK244 pKa = 10.97PHH246 pKa = 6.54DD247 pKa = 3.56VRR249 pKa = 11.84LGGRR253 pKa = 11.84GKK255 pKa = 9.95VAEE258 pKa = 4.32MDD260 pKa = 3.35RR261 pKa = 11.84TAAEE265 pKa = 4.26AKK267 pKa = 9.31PPAVGRR273 pKa = 11.84LILMMSQRR281 pKa = 11.84DD282 pKa = 3.76LLLCGVTEE290 pKa = 4.19NLLTRR295 pKa = 11.84AYY297 pKa = 9.36TPAQFPIAVGQSWFHH312 pKa = 6.17GGATEE317 pKa = 4.45FVDD320 pKa = 3.96RR321 pKa = 11.84FYY323 pKa = 10.71PYY325 pKa = 10.19TEE327 pKa = 4.27YY328 pKa = 11.15FCFDD332 pKa = 3.16AKK334 pKa = 10.95KK335 pKa = 9.98FDD337 pKa = 3.75SSIDD341 pKa = 2.94RR342 pKa = 11.84WMVRR346 pKa = 11.84IAINILRR353 pKa = 11.84HH354 pKa = 4.61QFFEE358 pKa = 4.64GEE360 pKa = 3.95EE361 pKa = 4.23SKK363 pKa = 11.47YY364 pKa = 10.22DD365 pKa = 3.65AYY367 pKa = 8.98WTFVEE372 pKa = 5.19EE373 pKa = 4.48SLLDD377 pKa = 3.59APIYY381 pKa = 10.49RR382 pKa = 11.84DD383 pKa = 3.52DD384 pKa = 3.58GVRR387 pKa = 11.84FQKK390 pKa = 10.75HH391 pKa = 5.89CGTTSGHH398 pKa = 5.12SHH400 pKa = 5.6NTLIQSIITLLLAYY414 pKa = 7.21TTMLFLHH421 pKa = 7.07PEE423 pKa = 4.14LSVEE427 pKa = 4.37EE428 pKa = 4.15ILEE431 pKa = 4.17NMWAEE436 pKa = 4.24SLGDD440 pKa = 3.91DD441 pKa = 5.08NILGVKK447 pKa = 10.2GPLAGHH453 pKa = 5.33TVEE456 pKa = 5.9EE457 pKa = 4.48IAQVVYY463 pKa = 10.85DD464 pKa = 4.02IFRR467 pKa = 11.84VDD469 pKa = 3.0WFGKK473 pKa = 10.21KK474 pKa = 8.56SFKK477 pKa = 7.89TTCLLDD483 pKa = 4.81AITGAFQGLQFLGKK497 pKa = 8.47YY498 pKa = 9.33FRR500 pKa = 11.84IGEE503 pKa = 4.26YY504 pKa = 8.29PTADD508 pKa = 3.89KK509 pKa = 10.9LVDD512 pKa = 3.43MPIPYY517 pKa = 10.14RR518 pKa = 11.84PFKK521 pKa = 9.52EE522 pKa = 4.21TYY524 pKa = 10.02LRR526 pKa = 11.84LLYY529 pKa = 9.91PEE531 pKa = 4.91YY532 pKa = 10.53GAHH535 pKa = 6.12TPTEE539 pKa = 3.78TWLRR543 pKa = 11.84TLGNYY548 pKa = 9.75LDD550 pKa = 4.58AAGNPQAEE558 pKa = 4.49TWLSGLLTWLEE569 pKa = 4.33DD570 pKa = 3.63RR571 pKa = 11.84VEE573 pKa = 4.34TPPEE577 pKa = 3.79EE578 pKa = 3.91WPANYY583 pKa = 9.92KK584 pKa = 10.68RR585 pKa = 11.84MVSRR589 pKa = 11.84DD590 pKa = 3.52YY591 pKa = 10.61SHH593 pKa = 7.85VGIEE597 pKa = 4.03VPKK600 pKa = 9.78PVRR603 pKa = 11.84INYY606 pKa = 6.99EE607 pKa = 3.45QWRR610 pKa = 11.84DD611 pKa = 3.53LVVMSRR617 pKa = 11.84ADD619 pKa = 3.39YY620 pKa = 11.13VNTYY624 pKa = 10.22KK625 pKa = 10.74PALVKK630 pKa = 10.49QQ631 pKa = 3.73
MM1 pKa = 7.49NAPEE5 pKa = 4.12IPGRR9 pKa = 11.84NPDD12 pKa = 3.83RR13 pKa = 11.84IRR15 pKa = 11.84RR16 pKa = 11.84SSLFNFDD23 pKa = 4.26GGLLTDD29 pKa = 3.2SHH31 pKa = 8.25GDD33 pKa = 3.31VLLNTRR39 pKa = 11.84KK40 pKa = 10.04RR41 pKa = 11.84GRR43 pKa = 11.84NQFDD47 pKa = 4.42DD48 pKa = 3.93EE49 pKa = 4.88TEE51 pKa = 4.26VTSAPSSHH59 pKa = 6.43TSGNASTRR67 pKa = 11.84RR68 pKa = 11.84RR69 pKa = 11.84KK70 pKa = 9.35RR71 pKa = 11.84LRR73 pKa = 11.84RR74 pKa = 11.84VRR76 pKa = 11.84MASHH80 pKa = 6.35HH81 pKa = 5.6TDD83 pKa = 2.95EE84 pKa = 4.8MSALTKK90 pKa = 10.41LFQHH94 pKa = 6.66ISILGRR100 pKa = 11.84IEE102 pKa = 4.09QYY104 pKa = 11.38NKK106 pKa = 10.49LNPDD110 pKa = 3.97LVVADD115 pKa = 4.66PDD117 pKa = 4.13PLIVEE122 pKa = 4.56YY123 pKa = 9.84TLHH126 pKa = 6.48NPPIIPNFKK135 pKa = 10.02PEE137 pKa = 4.74DD138 pKa = 3.62YY139 pKa = 10.18TFVKK143 pKa = 10.58ACTPVEE149 pKa = 4.1MAHH152 pKa = 6.73LKK154 pKa = 10.74HH155 pKa = 6.62FDD157 pKa = 3.59RR158 pKa = 11.84EE159 pKa = 4.18EE160 pKa = 4.14SVPQAEE166 pKa = 4.75YY167 pKa = 10.9LPSLQTASDD176 pKa = 3.46IVGKK180 pKa = 10.17LLSLPEE186 pKa = 3.94YY187 pKa = 10.87LFFPEE192 pKa = 5.42VDD194 pKa = 3.45DD195 pKa = 4.73LNNVRR200 pKa = 11.84YY201 pKa = 9.68IGSKK205 pKa = 10.41FPGIEE210 pKa = 3.76YY211 pKa = 10.72AMQGFKK217 pKa = 9.43TRR219 pKa = 11.84RR220 pKa = 11.84EE221 pKa = 4.04AHH223 pKa = 5.73EE224 pKa = 3.99MAMTDD229 pKa = 3.52ALSAFDD235 pKa = 4.29KK236 pKa = 10.76LLNGEE241 pKa = 4.19RR242 pKa = 11.84VKK244 pKa = 10.97PHH246 pKa = 6.54DD247 pKa = 3.56VRR249 pKa = 11.84LGGRR253 pKa = 11.84GKK255 pKa = 9.95VAEE258 pKa = 4.32MDD260 pKa = 3.35RR261 pKa = 11.84TAAEE265 pKa = 4.26AKK267 pKa = 9.31PPAVGRR273 pKa = 11.84LILMMSQRR281 pKa = 11.84DD282 pKa = 3.76LLLCGVTEE290 pKa = 4.19NLLTRR295 pKa = 11.84AYY297 pKa = 9.36TPAQFPIAVGQSWFHH312 pKa = 6.17GGATEE317 pKa = 4.45FVDD320 pKa = 3.96RR321 pKa = 11.84FYY323 pKa = 10.71PYY325 pKa = 10.19TEE327 pKa = 4.27YY328 pKa = 11.15FCFDD332 pKa = 3.16AKK334 pKa = 10.95KK335 pKa = 9.98FDD337 pKa = 3.75SSIDD341 pKa = 2.94RR342 pKa = 11.84WMVRR346 pKa = 11.84IAINILRR353 pKa = 11.84HH354 pKa = 4.61QFFEE358 pKa = 4.64GEE360 pKa = 3.95EE361 pKa = 4.23SKK363 pKa = 11.47YY364 pKa = 10.22DD365 pKa = 3.65AYY367 pKa = 8.98WTFVEE372 pKa = 5.19EE373 pKa = 4.48SLLDD377 pKa = 3.59APIYY381 pKa = 10.49RR382 pKa = 11.84DD383 pKa = 3.52DD384 pKa = 3.58GVRR387 pKa = 11.84FQKK390 pKa = 10.75HH391 pKa = 5.89CGTTSGHH398 pKa = 5.12SHH400 pKa = 5.6NTLIQSIITLLLAYY414 pKa = 7.21TTMLFLHH421 pKa = 7.07PEE423 pKa = 4.14LSVEE427 pKa = 4.37EE428 pKa = 4.15ILEE431 pKa = 4.17NMWAEE436 pKa = 4.24SLGDD440 pKa = 3.91DD441 pKa = 5.08NILGVKK447 pKa = 10.2GPLAGHH453 pKa = 5.33TVEE456 pKa = 5.9EE457 pKa = 4.48IAQVVYY463 pKa = 10.85DD464 pKa = 4.02IFRR467 pKa = 11.84VDD469 pKa = 3.0WFGKK473 pKa = 10.21KK474 pKa = 8.56SFKK477 pKa = 7.89TTCLLDD483 pKa = 4.81AITGAFQGLQFLGKK497 pKa = 8.47YY498 pKa = 9.33FRR500 pKa = 11.84IGEE503 pKa = 4.26YY504 pKa = 8.29PTADD508 pKa = 3.89KK509 pKa = 10.9LVDD512 pKa = 3.43MPIPYY517 pKa = 10.14RR518 pKa = 11.84PFKK521 pKa = 9.52EE522 pKa = 4.21TYY524 pKa = 10.02LRR526 pKa = 11.84LLYY529 pKa = 9.91PEE531 pKa = 4.91YY532 pKa = 10.53GAHH535 pKa = 6.12TPTEE539 pKa = 3.78TWLRR543 pKa = 11.84TLGNYY548 pKa = 9.75LDD550 pKa = 4.58AAGNPQAEE558 pKa = 4.49TWLSGLLTWLEE569 pKa = 4.33DD570 pKa = 3.63RR571 pKa = 11.84VEE573 pKa = 4.34TPPEE577 pKa = 3.79EE578 pKa = 3.91WPANYY583 pKa = 9.92KK584 pKa = 10.68RR585 pKa = 11.84MVSRR589 pKa = 11.84DD590 pKa = 3.52YY591 pKa = 10.61SHH593 pKa = 7.85VGIEE597 pKa = 4.03VPKK600 pKa = 9.78PVRR603 pKa = 11.84INYY606 pKa = 6.99EE607 pKa = 3.45QWRR610 pKa = 11.84DD611 pKa = 3.53LVVMSRR617 pKa = 11.84ADD619 pKa = 3.39YY620 pKa = 11.13VNTYY624 pKa = 10.22KK625 pKa = 10.74PALVKK630 pKa = 10.49QQ631 pKa = 3.73
Molecular weight: 72.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A346THI6|A0A346THI6_9VIRU Uncharacterized protein OS=Trichoderma harzianum bipartite mycovirus 1 OX=2315392 PE=4 SV=1
MM1 pKa = 7.7AAQPQPSLLAQFGGANTNIDD21 pKa = 4.11FSADD25 pKa = 3.59PNRR28 pKa = 11.84LAAALPIPEE37 pKa = 4.75MGDD40 pKa = 3.26SFEE43 pKa = 3.98EE44 pKa = 4.12LQRR47 pKa = 11.84KK48 pKa = 8.21LRR50 pKa = 11.84ALAHH54 pKa = 5.4VVEE57 pKa = 4.82QGRR60 pKa = 11.84SVAVATGYY68 pKa = 10.24KK69 pKa = 9.99AAAQQEE75 pKa = 4.16RR76 pKa = 11.84DD77 pKa = 3.44PRR79 pKa = 11.84EE80 pKa = 3.88IARR83 pKa = 11.84MAMTPVEE90 pKa = 4.31IMQLDD95 pKa = 3.41AWEE98 pKa = 4.47TGAALPKK105 pKa = 9.73MDD107 pKa = 3.29WGKK110 pKa = 10.22DD111 pKa = 3.27QGEE114 pKa = 4.3PPQGDD119 pKa = 3.55ASRR122 pKa = 11.84ATALRR127 pKa = 11.84RR128 pKa = 11.84AAALNRR134 pKa = 11.84LYY136 pKa = 9.8KK137 pKa = 9.56TDD139 pKa = 3.36KK140 pKa = 10.52ATVEE144 pKa = 3.71NSTYY148 pKa = 9.98FVKK151 pKa = 10.63NYY153 pKa = 10.72SPLIPLLTAVTKK165 pKa = 10.58IIGAQRR171 pKa = 11.84DD172 pKa = 3.76LVGGMEE178 pKa = 4.86EE179 pKa = 4.19LDD181 pKa = 3.71EE182 pKa = 4.09MQMAEE187 pKa = 3.86IQTIGKK193 pKa = 9.02IVATAQTNINRR204 pKa = 11.84IQEE207 pKa = 4.06EE208 pKa = 4.35VRR210 pKa = 11.84RR211 pKa = 11.84LLRR214 pKa = 11.84DD215 pKa = 3.18INYY218 pKa = 10.18SNSVLLARR226 pKa = 11.84SHH228 pKa = 6.41ALKK231 pKa = 10.63EE232 pKa = 4.31LIPKK236 pKa = 9.18YY237 pKa = 10.0KK238 pKa = 10.14KK239 pKa = 10.63KK240 pKa = 10.38KK241 pKa = 10.55DD242 pKa = 3.66PVNDD246 pKa = 2.85ARR248 pKa = 11.84LAVGRR253 pKa = 11.84PAIGRR258 pKa = 11.84SFDD261 pKa = 3.71YY262 pKa = 10.42RR263 pKa = 11.84SKK265 pKa = 11.02AAGARR270 pKa = 11.84EE271 pKa = 3.74DD272 pKa = 3.32RR273 pKa = 11.84TNFLRR278 pKa = 11.84GTGLVAGRR286 pKa = 11.84AGPYY290 pKa = 8.86PRR292 pKa = 11.84EE293 pKa = 4.31AKK295 pKa = 9.84RR296 pKa = 11.84VRR298 pKa = 11.84TQSPNPPAEE307 pKa = 4.36EE308 pKa = 4.63GEE310 pKa = 4.31PMAQQ314 pKa = 3.28
MM1 pKa = 7.7AAQPQPSLLAQFGGANTNIDD21 pKa = 4.11FSADD25 pKa = 3.59PNRR28 pKa = 11.84LAAALPIPEE37 pKa = 4.75MGDD40 pKa = 3.26SFEE43 pKa = 3.98EE44 pKa = 4.12LQRR47 pKa = 11.84KK48 pKa = 8.21LRR50 pKa = 11.84ALAHH54 pKa = 5.4VVEE57 pKa = 4.82QGRR60 pKa = 11.84SVAVATGYY68 pKa = 10.24KK69 pKa = 9.99AAAQQEE75 pKa = 4.16RR76 pKa = 11.84DD77 pKa = 3.44PRR79 pKa = 11.84EE80 pKa = 3.88IARR83 pKa = 11.84MAMTPVEE90 pKa = 4.31IMQLDD95 pKa = 3.41AWEE98 pKa = 4.47TGAALPKK105 pKa = 9.73MDD107 pKa = 3.29WGKK110 pKa = 10.22DD111 pKa = 3.27QGEE114 pKa = 4.3PPQGDD119 pKa = 3.55ASRR122 pKa = 11.84ATALRR127 pKa = 11.84RR128 pKa = 11.84AAALNRR134 pKa = 11.84LYY136 pKa = 9.8KK137 pKa = 9.56TDD139 pKa = 3.36KK140 pKa = 10.52ATVEE144 pKa = 3.71NSTYY148 pKa = 9.98FVKK151 pKa = 10.63NYY153 pKa = 10.72SPLIPLLTAVTKK165 pKa = 10.58IIGAQRR171 pKa = 11.84DD172 pKa = 3.76LVGGMEE178 pKa = 4.86EE179 pKa = 4.19LDD181 pKa = 3.71EE182 pKa = 4.09MQMAEE187 pKa = 3.86IQTIGKK193 pKa = 9.02IVATAQTNINRR204 pKa = 11.84IQEE207 pKa = 4.06EE208 pKa = 4.35VRR210 pKa = 11.84RR211 pKa = 11.84LLRR214 pKa = 11.84DD215 pKa = 3.18INYY218 pKa = 10.18SNSVLLARR226 pKa = 11.84SHH228 pKa = 6.41ALKK231 pKa = 10.63EE232 pKa = 4.31LIPKK236 pKa = 9.18YY237 pKa = 10.0KK238 pKa = 10.14KK239 pKa = 10.63KK240 pKa = 10.38KK241 pKa = 10.55DD242 pKa = 3.66PVNDD246 pKa = 2.85ARR248 pKa = 11.84LAVGRR253 pKa = 11.84PAIGRR258 pKa = 11.84SFDD261 pKa = 3.71YY262 pKa = 10.42RR263 pKa = 11.84SKK265 pKa = 11.02AAGARR270 pKa = 11.84EE271 pKa = 3.74DD272 pKa = 3.32RR273 pKa = 11.84TNFLRR278 pKa = 11.84GTGLVAGRR286 pKa = 11.84AGPYY290 pKa = 8.86PRR292 pKa = 11.84EE293 pKa = 4.31AKK295 pKa = 9.84RR296 pKa = 11.84VRR298 pKa = 11.84TQSPNPPAEE307 pKa = 4.36EE308 pKa = 4.63GEE310 pKa = 4.31PMAQQ314 pKa = 3.28
Molecular weight: 34.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
945 |
314 |
631 |
472.5 |
53.37 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.206 ± 3.021 | 0.529 ± 0.312 |
5.926 ± 0.49 | 7.09 ± 0.049 |
3.915 ± 1.182 | 6.243 ± 0.262 |
2.222 ± 0.935 | 4.762 ± 0.009 |
4.868 ± 0.322 | 9.418 ± 0.671 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.256 | 3.81 ± 0.195 |
6.349 ± 0.2 | 3.704 ± 1.008 |
7.513 ± 0.828 | 4.868 ± 0.429 |
6.032 ± 0.552 | 5.714 ± 0.365 |
1.27 ± 0.373 | 3.81 ± 0.744 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
