
Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H) (Methanobacterium thermoautotrophicum)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Methanomada group; Methanobacteria; Methanobacteriales; Methanobacteriaceae; Methanothermobacter; Methanothermobacter thermautotrophicus
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
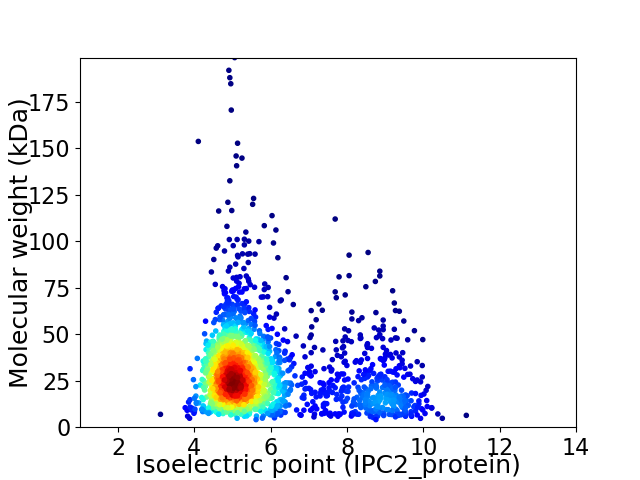
Virtual 2D-PAGE plot for 1869 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|O26563|O26563_METTH Uncharacterized protein OS=Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H) OX=187420 GN=MTH_463 PE=3 SV=1
MM1 pKa = 7.82SDD3 pKa = 4.05IDD5 pKa = 5.77DD6 pKa = 3.77EE7 pKa = 4.59TADD10 pKa = 3.66VLVEE14 pKa = 3.84LVNEE18 pKa = 4.16YY19 pKa = 10.43EE20 pKa = 4.19YY21 pKa = 11.48EE22 pKa = 4.03HH23 pKa = 7.81LLDD26 pKa = 4.73KK27 pKa = 11.2EE28 pKa = 4.55SVNDD32 pKa = 3.28GWKK35 pKa = 10.24RR36 pKa = 11.84FTEE39 pKa = 4.59AFYY42 pKa = 11.24DD43 pKa = 3.84DD44 pKa = 4.48NKK46 pKa = 11.31EE47 pKa = 4.14MLCDD51 pKa = 3.21TAFNYY56 pKa = 10.48VIDD59 pKa = 4.42GMPDD63 pKa = 3.06DD64 pKa = 5.65GIRR67 pKa = 11.84EE68 pKa = 4.38VFSRR72 pKa = 11.84TYY74 pKa = 10.68LDD76 pKa = 3.11NRR78 pKa = 11.84EE79 pKa = 4.12YY80 pKa = 10.4IISSFRR86 pKa = 11.84YY87 pKa = 9.8DD88 pKa = 3.49EE89 pKa = 5.4DD90 pKa = 4.35FIAYY94 pKa = 9.78FDD96 pKa = 4.51ADD98 pKa = 4.34YY99 pKa = 11.12MDD101 pKa = 6.23LDD103 pKa = 4.63LSNVDD108 pKa = 5.41LGDD111 pKa = 4.19FDD113 pKa = 5.3PLMVRR118 pKa = 11.84SWFIDD123 pKa = 3.06MLLPVALYY131 pKa = 8.36ITEE134 pKa = 4.01VMVDD138 pKa = 4.02FSYY141 pKa = 11.41DD142 pKa = 3.41LEE144 pKa = 4.88SYY146 pKa = 10.52CSVYY150 pKa = 10.56TFTVEE155 pKa = 3.92DD156 pKa = 4.35GEE158 pKa = 4.44IVDD161 pKa = 3.82VEE163 pKa = 4.28FNEE166 pKa = 4.37RR167 pKa = 11.84FIDD170 pKa = 4.03GEE172 pKa = 4.33GMMDD176 pKa = 4.05LSEE179 pKa = 4.69HH180 pKa = 6.16SLLEE184 pKa = 4.13KK185 pKa = 10.43LDD187 pKa = 4.41EE188 pKa = 4.58IKK190 pKa = 11.19NPFDD194 pKa = 3.24MNRR197 pKa = 11.84RR198 pKa = 11.84MRR200 pKa = 11.84KK201 pKa = 8.67ICSTDD206 pKa = 2.89VGRR209 pKa = 11.84EE210 pKa = 3.41EE211 pKa = 4.58ALRR214 pKa = 11.84MSYY217 pKa = 10.57FFDD220 pKa = 4.91CVDD223 pKa = 4.16CEE225 pKa = 4.47VKK227 pKa = 10.69VDD229 pKa = 3.41ILKK232 pKa = 8.72YY233 pKa = 9.53TFDD236 pKa = 4.0FLCRR240 pKa = 11.84LAPDD244 pKa = 3.71EE245 pKa = 4.09TASYY249 pKa = 9.67YY250 pKa = 9.83ISKK253 pKa = 10.51ARR255 pKa = 11.84DD256 pKa = 3.27HH257 pKa = 6.31MFEE260 pKa = 4.32IYY262 pKa = 10.27QIEE265 pKa = 3.92
MM1 pKa = 7.82SDD3 pKa = 4.05IDD5 pKa = 5.77DD6 pKa = 3.77EE7 pKa = 4.59TADD10 pKa = 3.66VLVEE14 pKa = 3.84LVNEE18 pKa = 4.16YY19 pKa = 10.43EE20 pKa = 4.19YY21 pKa = 11.48EE22 pKa = 4.03HH23 pKa = 7.81LLDD26 pKa = 4.73KK27 pKa = 11.2EE28 pKa = 4.55SVNDD32 pKa = 3.28GWKK35 pKa = 10.24RR36 pKa = 11.84FTEE39 pKa = 4.59AFYY42 pKa = 11.24DD43 pKa = 3.84DD44 pKa = 4.48NKK46 pKa = 11.31EE47 pKa = 4.14MLCDD51 pKa = 3.21TAFNYY56 pKa = 10.48VIDD59 pKa = 4.42GMPDD63 pKa = 3.06DD64 pKa = 5.65GIRR67 pKa = 11.84EE68 pKa = 4.38VFSRR72 pKa = 11.84TYY74 pKa = 10.68LDD76 pKa = 3.11NRR78 pKa = 11.84EE79 pKa = 4.12YY80 pKa = 10.4IISSFRR86 pKa = 11.84YY87 pKa = 9.8DD88 pKa = 3.49EE89 pKa = 5.4DD90 pKa = 4.35FIAYY94 pKa = 9.78FDD96 pKa = 4.51ADD98 pKa = 4.34YY99 pKa = 11.12MDD101 pKa = 6.23LDD103 pKa = 4.63LSNVDD108 pKa = 5.41LGDD111 pKa = 4.19FDD113 pKa = 5.3PLMVRR118 pKa = 11.84SWFIDD123 pKa = 3.06MLLPVALYY131 pKa = 8.36ITEE134 pKa = 4.01VMVDD138 pKa = 4.02FSYY141 pKa = 11.41DD142 pKa = 3.41LEE144 pKa = 4.88SYY146 pKa = 10.52CSVYY150 pKa = 10.56TFTVEE155 pKa = 3.92DD156 pKa = 4.35GEE158 pKa = 4.44IVDD161 pKa = 3.82VEE163 pKa = 4.28FNEE166 pKa = 4.37RR167 pKa = 11.84FIDD170 pKa = 4.03GEE172 pKa = 4.33GMMDD176 pKa = 4.05LSEE179 pKa = 4.69HH180 pKa = 6.16SLLEE184 pKa = 4.13KK185 pKa = 10.43LDD187 pKa = 4.41EE188 pKa = 4.58IKK190 pKa = 11.19NPFDD194 pKa = 3.24MNRR197 pKa = 11.84RR198 pKa = 11.84MRR200 pKa = 11.84KK201 pKa = 8.67ICSTDD206 pKa = 2.89VGRR209 pKa = 11.84EE210 pKa = 3.41EE211 pKa = 4.58ALRR214 pKa = 11.84MSYY217 pKa = 10.57FFDD220 pKa = 4.91CVDD223 pKa = 4.16CEE225 pKa = 4.47VKK227 pKa = 10.69VDD229 pKa = 3.41ILKK232 pKa = 8.72YY233 pKa = 9.53TFDD236 pKa = 4.0FLCRR240 pKa = 11.84LAPDD244 pKa = 3.71EE245 pKa = 4.09TASYY249 pKa = 9.67YY250 pKa = 9.83ISKK253 pKa = 10.51ARR255 pKa = 11.84DD256 pKa = 3.27HH257 pKa = 6.31MFEE260 pKa = 4.32IYY262 pKa = 10.27QIEE265 pKa = 3.92
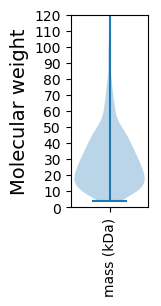
Molecular weight: 31.46 kDa
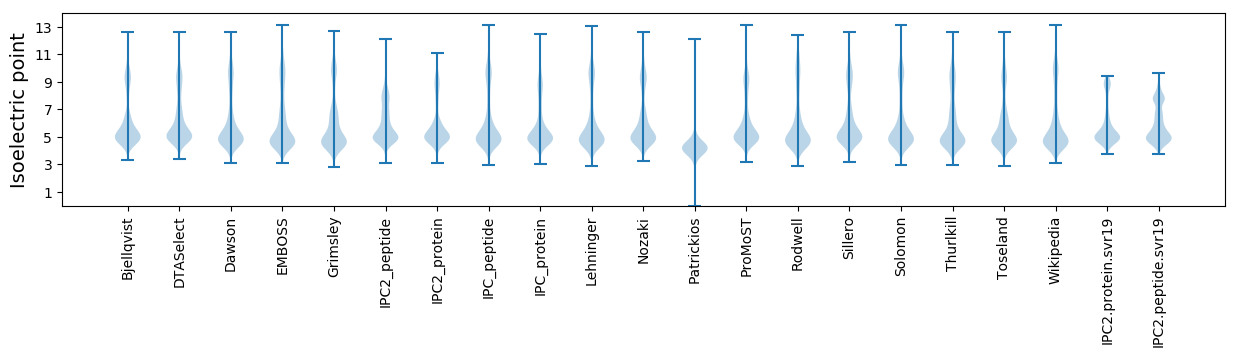
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O27654|O27654_METTH Conserved protein OS=Methanothermobacter thermautotrophicus (strain ATCC 29096 / DSM 1053 / JCM 10044 / NBRC 100330 / Delta H) OX=187420 GN=MTH_1617 PE=4 SV=1
MM1 pKa = 7.39SRR3 pKa = 11.84NKK5 pKa = 10.04HH6 pKa = 4.23VARR9 pKa = 11.84KK10 pKa = 9.84LRR12 pKa = 11.84MAKK15 pKa = 10.25ANRR18 pKa = 11.84QNRR21 pKa = 11.84RR22 pKa = 11.84VPAWVMVKK30 pKa = 9.15TNYY33 pKa = 9.7RR34 pKa = 11.84VRR36 pKa = 11.84SHH38 pKa = 6.66PKK40 pKa = 7.08MRR42 pKa = 11.84HH43 pKa = 3.26WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.85LKK50 pKa = 10.03VV51 pKa = 2.95
MM1 pKa = 7.39SRR3 pKa = 11.84NKK5 pKa = 10.04HH6 pKa = 4.23VARR9 pKa = 11.84KK10 pKa = 9.84LRR12 pKa = 11.84MAKK15 pKa = 10.25ANRR18 pKa = 11.84QNRR21 pKa = 11.84RR22 pKa = 11.84VPAWVMVKK30 pKa = 9.15TNYY33 pKa = 9.7RR34 pKa = 11.84VRR36 pKa = 11.84SHH38 pKa = 6.66PKK40 pKa = 7.08MRR42 pKa = 11.84HH43 pKa = 3.26WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.85LKK50 pKa = 10.03VV51 pKa = 2.95
Molecular weight: 6.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
524739 |
35 |
1787 |
280.8 |
31.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.342 ± 0.058 | 1.208 ± 0.032 |
5.914 ± 0.04 | 8.141 ± 0.066 |
3.648 ± 0.039 | 7.979 ± 0.055 |
1.874 ± 0.024 | 7.711 ± 0.045 |
4.558 ± 0.06 | 9.425 ± 0.07 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.061 ± 0.027 | 3.309 ± 0.06 |
4.3 ± 0.033 | 1.895 ± 0.026 |
6.796 ± 0.062 | 6.14 ± 0.044 |
4.962 ± 0.054 | 7.674 ± 0.055 |
0.84 ± 0.018 | 3.224 ± 0.04 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
