
Odonata associated gemycircularvirus-2
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus odona2; Odonata associated gemycircularvirus 2
Average proteome isoelectric point is 7.44
Get precalculated fractions of proteins
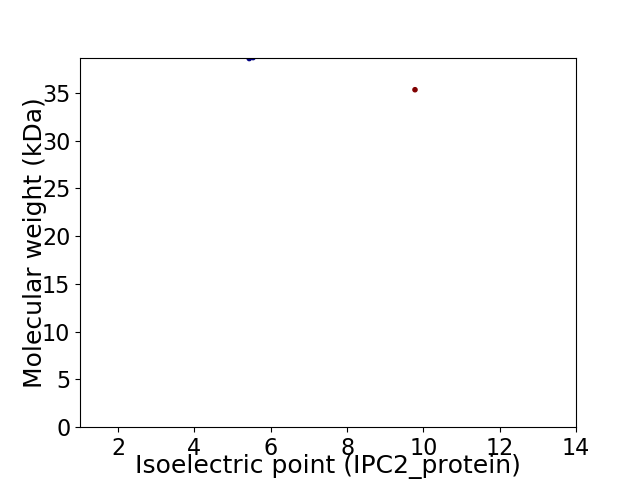
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
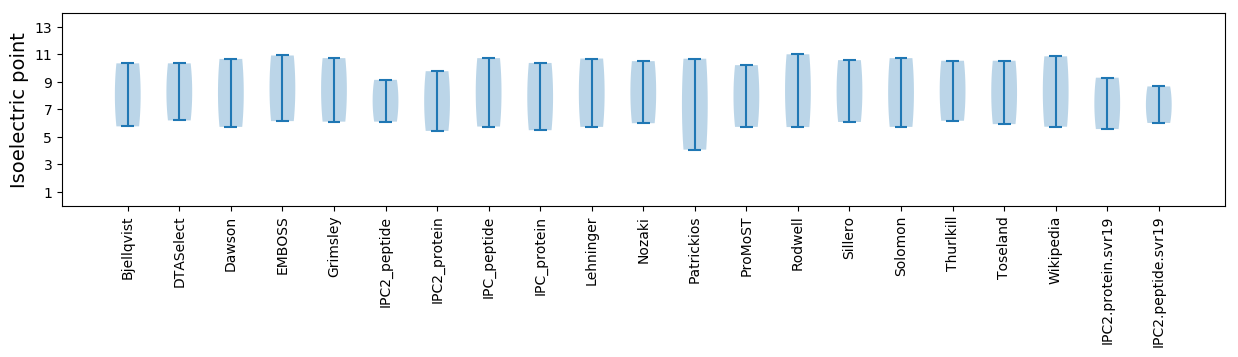
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B4UH04|A0A0B4UH04_9VIRU Replication-associated protein OS=Odonata associated gemycircularvirus-2 OX=1592108 PE=3 SV=1
MM1 pKa = 7.86APFHH5 pKa = 6.32LKK7 pKa = 9.3NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.35VLLTYY16 pKa = 10.3SQAGSEE22 pKa = 4.15FNYY25 pKa = 9.28WAIVDD30 pKa = 3.92MLSSHH35 pKa = 6.16GAEE38 pKa = 4.21CIIGRR43 pKa = 11.84EE44 pKa = 3.97LDD46 pKa = 3.55ADD48 pKa = 4.1GGTHH52 pKa = 5.57FHH54 pKa = 6.24VFVDD58 pKa = 4.51FGRR61 pKa = 11.84LFSTRR66 pKa = 11.84KK67 pKa = 8.19TNVFDD72 pKa = 4.58VDD74 pKa = 3.52GHH76 pKa = 6.54HH77 pKa = 7.3PNILPVWKK85 pKa = 8.83TPGEE89 pKa = 4.17AFDD92 pKa = 4.29YY93 pKa = 10.46AAKK96 pKa = 10.85DD97 pKa = 3.46GDD99 pKa = 3.89IVAGGLEE106 pKa = 4.42RR107 pKa = 11.84PGTDD111 pKa = 2.86CDD113 pKa = 3.8YY114 pKa = 11.07DD115 pKa = 3.71IEE117 pKa = 4.87NFWACAGASQSGEE130 pKa = 4.12EE131 pKa = 4.25FLHH134 pKa = 7.05FLDD137 pKa = 4.3QLAPRR142 pKa = 11.84DD143 pKa = 3.71LMRR146 pKa = 11.84GFIQFRR152 pKa = 11.84SYY154 pKa = 11.73ADD156 pKa = 3.42WKK158 pKa = 9.37WAVAPEE164 pKa = 4.48RR165 pKa = 11.84YY166 pKa = 9.68VNPPGVMFDD175 pKa = 3.49TGHH178 pKa = 7.26AEE180 pKa = 4.63QLSEE184 pKa = 4.08WLSQANLGSGPGRR197 pKa = 11.84VRR199 pKa = 11.84LVAHH203 pKa = 6.22LHH205 pKa = 5.95GSRR208 pKa = 11.84KK209 pKa = 9.99SLMLWGPTQYY219 pKa = 11.46GKK221 pKa = 6.97TTWARR226 pKa = 11.84SLGNHH231 pKa = 6.2IFFGSQFSGKK241 pKa = 9.72LALDD245 pKa = 4.14GMQDD249 pKa = 2.71AEE251 pKa = 4.41YY252 pKa = 10.96AVFDD256 pKa = 3.84DD257 pKa = 4.1WKK259 pKa = 10.9GGMKK263 pKa = 10.47ALPGYY268 pKa = 9.93KK269 pKa = 9.62DD270 pKa = 2.93WFGCQWQISVRR281 pKa = 11.84KK282 pKa = 8.73LHH284 pKa = 7.02HH285 pKa = 6.67DD286 pKa = 3.36AKK288 pKa = 10.63LITWGRR294 pKa = 11.84PIIWLCNKK302 pKa = 9.85DD303 pKa = 3.55PRR305 pKa = 11.84LMHH308 pKa = 6.42VATDD312 pKa = 3.78DD313 pKa = 4.83VDD315 pKa = 3.98WEE317 pKa = 4.22WMDD320 pKa = 4.21DD321 pKa = 3.37NVIFVEE327 pKa = 4.38LARR330 pKa = 11.84PLATFRR336 pKa = 11.84ASTEE340 pKa = 3.78
MM1 pKa = 7.86APFHH5 pKa = 6.32LKK7 pKa = 9.3NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.35VLLTYY16 pKa = 10.3SQAGSEE22 pKa = 4.15FNYY25 pKa = 9.28WAIVDD30 pKa = 3.92MLSSHH35 pKa = 6.16GAEE38 pKa = 4.21CIIGRR43 pKa = 11.84EE44 pKa = 3.97LDD46 pKa = 3.55ADD48 pKa = 4.1GGTHH52 pKa = 5.57FHH54 pKa = 6.24VFVDD58 pKa = 4.51FGRR61 pKa = 11.84LFSTRR66 pKa = 11.84KK67 pKa = 8.19TNVFDD72 pKa = 4.58VDD74 pKa = 3.52GHH76 pKa = 6.54HH77 pKa = 7.3PNILPVWKK85 pKa = 8.83TPGEE89 pKa = 4.17AFDD92 pKa = 4.29YY93 pKa = 10.46AAKK96 pKa = 10.85DD97 pKa = 3.46GDD99 pKa = 3.89IVAGGLEE106 pKa = 4.42RR107 pKa = 11.84PGTDD111 pKa = 2.86CDD113 pKa = 3.8YY114 pKa = 11.07DD115 pKa = 3.71IEE117 pKa = 4.87NFWACAGASQSGEE130 pKa = 4.12EE131 pKa = 4.25FLHH134 pKa = 7.05FLDD137 pKa = 4.3QLAPRR142 pKa = 11.84DD143 pKa = 3.71LMRR146 pKa = 11.84GFIQFRR152 pKa = 11.84SYY154 pKa = 11.73ADD156 pKa = 3.42WKK158 pKa = 9.37WAVAPEE164 pKa = 4.48RR165 pKa = 11.84YY166 pKa = 9.68VNPPGVMFDD175 pKa = 3.49TGHH178 pKa = 7.26AEE180 pKa = 4.63QLSEE184 pKa = 4.08WLSQANLGSGPGRR197 pKa = 11.84VRR199 pKa = 11.84LVAHH203 pKa = 6.22LHH205 pKa = 5.95GSRR208 pKa = 11.84KK209 pKa = 9.99SLMLWGPTQYY219 pKa = 11.46GKK221 pKa = 6.97TTWARR226 pKa = 11.84SLGNHH231 pKa = 6.2IFFGSQFSGKK241 pKa = 9.72LALDD245 pKa = 4.14GMQDD249 pKa = 2.71AEE251 pKa = 4.41YY252 pKa = 10.96AVFDD256 pKa = 3.84DD257 pKa = 4.1WKK259 pKa = 10.9GGMKK263 pKa = 10.47ALPGYY268 pKa = 9.93KK269 pKa = 9.62DD270 pKa = 2.93WFGCQWQISVRR281 pKa = 11.84KK282 pKa = 8.73LHH284 pKa = 7.02HH285 pKa = 6.67DD286 pKa = 3.36AKK288 pKa = 10.63LITWGRR294 pKa = 11.84PIIWLCNKK302 pKa = 9.85DD303 pKa = 3.55PRR305 pKa = 11.84LMHH308 pKa = 6.42VATDD312 pKa = 3.78DD313 pKa = 4.83VDD315 pKa = 3.98WEE317 pKa = 4.22WMDD320 pKa = 4.21DD321 pKa = 3.37NVIFVEE327 pKa = 4.38LARR330 pKa = 11.84PLATFRR336 pKa = 11.84ASTEE340 pKa = 3.78
Molecular weight: 38.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B4UH04|A0A0B4UH04_9VIRU Replication-associated protein OS=Odonata associated gemycircularvirus-2 OX=1592108 PE=3 SV=1
MM1 pKa = 7.56VYY3 pKa = 10.08RR4 pKa = 11.84SKK6 pKa = 10.87SRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TTRR13 pKa = 11.84RR14 pKa = 11.84SSTRR18 pKa = 11.84KK19 pKa = 9.28RR20 pKa = 11.84PSTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.06GKK30 pKa = 9.72KK31 pKa = 9.44RR32 pKa = 11.84STFRR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84MSSKK43 pKa = 10.59RR44 pKa = 11.84ILNLTSKK51 pKa = 10.45KK52 pKa = 10.44KK53 pKa = 9.48QDD55 pKa = 3.37NMLSFSNTASDD66 pKa = 3.9GTPTTVAQGSLYY78 pKa = 10.34VAGVQSGATSNYY90 pKa = 9.23GMSIFCPTARR100 pKa = 11.84SLVTAGPPISRR111 pKa = 11.84WMSQTEE117 pKa = 4.12PPPHH121 pKa = 5.12VTLRR125 pKa = 11.84GYY127 pKa = 10.55KK128 pKa = 9.86EE129 pKa = 3.74NLRR132 pKa = 11.84IQTSSSQPWLWRR144 pKa = 11.84RR145 pKa = 11.84IVFNTKK151 pKa = 10.0GPTFSSTSVGDD162 pKa = 3.18TATRR166 pKa = 11.84QKK168 pKa = 10.92YY169 pKa = 10.21SPFSDD174 pKa = 3.35TSIGMARR181 pKa = 11.84LWFNTSVNAMSATQSLFQTVMFKK204 pKa = 10.99GAINQDD210 pKa = 2.59WSDD213 pKa = 4.48PITAKK218 pKa = 10.45VDD220 pKa = 3.34TSRR223 pKa = 11.84ITVMSDD229 pKa = 2.6RR230 pKa = 11.84TQTIKK235 pKa = 9.22TQNSNGHH242 pKa = 4.81FSEE245 pKa = 4.42RR246 pKa = 11.84KK247 pKa = 8.63LWYY250 pKa = 9.36PMKK253 pKa = 10.69KK254 pKa = 10.08NLVYY258 pKa = 10.94DD259 pKa = 4.23DD260 pKa = 5.4DD261 pKa = 4.16EE262 pKa = 6.9AGAAEE267 pKa = 4.31VPAYY271 pKa = 10.51YY272 pKa = 9.39STDD275 pKa = 3.29AKK277 pKa = 10.74PGMGDD282 pKa = 3.26VYY284 pKa = 10.82IVDD287 pKa = 4.46FVMPSVGATASDD299 pKa = 3.91IINFNCTSTLYY310 pKa = 9.52WHH312 pKa = 7.05EE313 pKa = 4.2KK314 pKa = 9.32
MM1 pKa = 7.56VYY3 pKa = 10.08RR4 pKa = 11.84SKK6 pKa = 10.87SRR8 pKa = 11.84RR9 pKa = 11.84RR10 pKa = 11.84TTRR13 pKa = 11.84RR14 pKa = 11.84SSTRR18 pKa = 11.84KK19 pKa = 9.28RR20 pKa = 11.84PSTRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.06GKK30 pKa = 9.72KK31 pKa = 9.44RR32 pKa = 11.84STFRR36 pKa = 11.84SRR38 pKa = 11.84RR39 pKa = 11.84MSSKK43 pKa = 10.59RR44 pKa = 11.84ILNLTSKK51 pKa = 10.45KK52 pKa = 10.44KK53 pKa = 9.48QDD55 pKa = 3.37NMLSFSNTASDD66 pKa = 3.9GTPTTVAQGSLYY78 pKa = 10.34VAGVQSGATSNYY90 pKa = 9.23GMSIFCPTARR100 pKa = 11.84SLVTAGPPISRR111 pKa = 11.84WMSQTEE117 pKa = 4.12PPPHH121 pKa = 5.12VTLRR125 pKa = 11.84GYY127 pKa = 10.55KK128 pKa = 9.86EE129 pKa = 3.74NLRR132 pKa = 11.84IQTSSSQPWLWRR144 pKa = 11.84RR145 pKa = 11.84IVFNTKK151 pKa = 10.0GPTFSSTSVGDD162 pKa = 3.18TATRR166 pKa = 11.84QKK168 pKa = 10.92YY169 pKa = 10.21SPFSDD174 pKa = 3.35TSIGMARR181 pKa = 11.84LWFNTSVNAMSATQSLFQTVMFKK204 pKa = 10.99GAINQDD210 pKa = 2.59WSDD213 pKa = 4.48PITAKK218 pKa = 10.45VDD220 pKa = 3.34TSRR223 pKa = 11.84ITVMSDD229 pKa = 2.6RR230 pKa = 11.84TQTIKK235 pKa = 9.22TQNSNGHH242 pKa = 4.81FSEE245 pKa = 4.42RR246 pKa = 11.84KK247 pKa = 8.63LWYY250 pKa = 9.36PMKK253 pKa = 10.69KK254 pKa = 10.08NLVYY258 pKa = 10.94DD259 pKa = 4.23DD260 pKa = 5.4DD261 pKa = 4.16EE262 pKa = 6.9AGAAEE267 pKa = 4.31VPAYY271 pKa = 10.51YY272 pKa = 9.39STDD275 pKa = 3.29AKK277 pKa = 10.74PGMGDD282 pKa = 3.26VYY284 pKa = 10.82IVDD287 pKa = 4.46FVMPSVGATASDD299 pKa = 3.91IINFNCTSTLYY310 pKa = 9.52WHH312 pKa = 7.05EE313 pKa = 4.2KK314 pKa = 9.32
Molecular weight: 35.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
654 |
314 |
340 |
327.0 |
36.95 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.339 ± 0.948 | 1.07 ± 0.319 |
6.575 ± 1.323 | 3.211 ± 0.957 |
5.046 ± 0.901 | 7.492 ± 1.529 |
2.599 ± 1.21 | 3.976 ± 0.121 |
5.046 ± 0.739 | 6.422 ± 1.679 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.211 ± 0.449 | 3.67 ± 0.58 |
4.74 ± 0.262 | 3.517 ± 0.224 |
7.034 ± 1.152 | 9.174 ± 3.091 |
7.492 ± 2.689 | 5.657 ± 0.055 |
3.364 ± 0.835 | 3.364 ± 0.337 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
