
Agrilus planipennis (Emerald ash borer) (Agrilus marcopoli)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Panarthropoda; Arthropoda; Mandibulata; Pancrustacea; Hexapoda; Insecta; Dicondylia; Pterygota; Neoptera; Endopterygota; Coleoptera; Polyphaga; Elateriformia;
Average proteome isoelectric point is 6.74
Get precalculated fractions of proteins
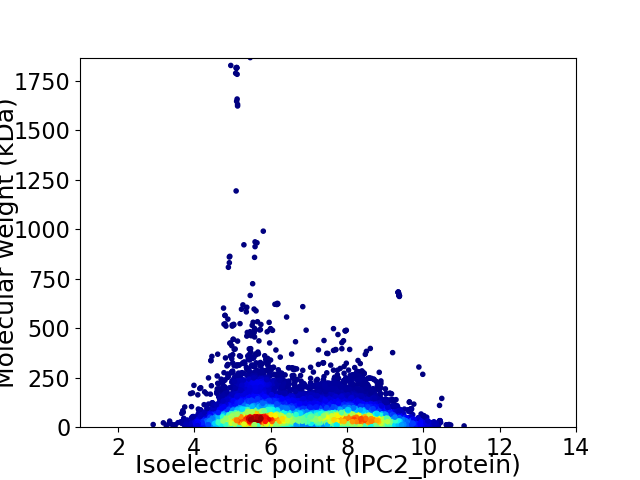
Virtual 2D-PAGE plot for 18772 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A7F5RLS0|A0A7F5RLS0_AGRPL apoptosis inhibitor 5 homolog OS=Agrilus planipennis OX=224129 GN=LOC112906595 PE=3 SV=1
MM1 pKa = 7.31NSLFKK6 pKa = 10.45FCAIVTVLLDD16 pKa = 3.87LSTYY20 pKa = 9.74NPRR23 pKa = 11.84GSNTYY28 pKa = 8.31NTDD31 pKa = 3.75RR32 pKa = 11.84IISTKK37 pKa = 9.68IDD39 pKa = 3.31AAQSILTTNLSFHH52 pKa = 7.03LATEE56 pKa = 4.32TTTLFSTTVSNTITSSTDD74 pKa = 3.02SFPDD78 pKa = 2.79RR79 pKa = 11.84STIVNTPEE87 pKa = 3.58THH89 pKa = 5.89LTTGSSEE96 pKa = 4.13EE97 pKa = 4.2SSTLNIIDD105 pKa = 5.39GYY107 pKa = 8.07TISYY111 pKa = 10.38APDD114 pKa = 3.8AEE116 pKa = 4.44LTTEE120 pKa = 4.03ITAEE124 pKa = 4.15TSATNAPDD132 pKa = 3.27RR133 pKa = 11.84STIPNRR139 pKa = 11.84PNTQLTTDD147 pKa = 3.41TTEE150 pKa = 3.77QSFTTNIPYY159 pKa = 10.7DD160 pKa = 3.6LTIPLSKK167 pKa = 10.83VKK169 pKa = 9.54LTTEE173 pKa = 4.76GIEE176 pKa = 4.01EE177 pKa = 5.55GYY179 pKa = 6.78TTTGYY184 pKa = 8.56STIPDD189 pKa = 3.59TSGTEE194 pKa = 3.97LPTEE198 pKa = 4.28STTGAITNTASNHH211 pKa = 4.47STLLSNSEE219 pKa = 4.17VEE221 pKa = 4.44LTTEE225 pKa = 4.07STTDD229 pKa = 3.18ASITNLSDD237 pKa = 3.14NLTISNTVGAEE248 pKa = 3.83IITEE252 pKa = 4.3GTVEE256 pKa = 4.05TSTTNVPNDD265 pKa = 3.56SASPNVPEE273 pKa = 4.09TSITSGTTQHH283 pKa = 6.94PFLTSIPDD291 pKa = 3.79DD292 pKa = 3.71STVSYY297 pKa = 10.39TPGAEE302 pKa = 3.98LTTEE306 pKa = 4.26GTKK309 pKa = 9.51DD310 pKa = 2.93TSTINLSDD318 pKa = 3.8NLTISSTTKK327 pKa = 10.67AKK329 pKa = 10.3IITEE333 pKa = 4.3GTVEE337 pKa = 4.05TSTTNVPNDD346 pKa = 3.56SASPNVPEE354 pKa = 4.31TSTTSGTTQHH364 pKa = 6.97PFLTSIPDD372 pKa = 3.79DD373 pKa = 3.71STVSYY378 pKa = 10.39TPGAEE383 pKa = 4.02LTTEE387 pKa = 4.27STTDD391 pKa = 2.96TYY393 pKa = 9.04TTNLSDD399 pKa = 4.87NLTIPNTAGAEE410 pKa = 4.03ITTEE414 pKa = 4.29GSAEE418 pKa = 4.01TSTTNVPKK426 pKa = 10.76DD427 pKa = 3.43STIPNAPEE435 pKa = 3.46IQMTLASTDD444 pKa = 3.32VHH446 pKa = 6.34STDD449 pKa = 3.64TDD451 pKa = 3.5HH452 pKa = 7.17GNSTISYY459 pKa = 7.93TPGVEE464 pKa = 3.96LTTEE468 pKa = 3.62ITTDD472 pKa = 3.1AFSTHH477 pKa = 6.87PSDD480 pKa = 5.23HH481 pKa = 5.99LTIPNTIEE489 pKa = 4.37AEE491 pKa = 3.78ITTEE495 pKa = 4.01VNGDD499 pKa = 3.6STISNMPVTEE509 pKa = 4.37LTTEE513 pKa = 4.12STTDD517 pKa = 2.99ASTTNLSDD525 pKa = 3.53NLTIPNTTGAEE536 pKa = 3.89ITTEE540 pKa = 4.27GSAEE544 pKa = 4.03TSTTNIPNDD553 pKa = 3.38STIPNISDD561 pKa = 2.81IQMALGSTDD570 pKa = 3.66EE571 pKa = 4.65PSTDD575 pKa = 3.44TVHH578 pKa = 6.9GDD580 pKa = 3.2STISYY585 pKa = 8.67TPGAEE590 pKa = 4.0LTTEE594 pKa = 4.19STTDD598 pKa = 2.86TSTINLSDD606 pKa = 3.68NLIIPNTTGAEE617 pKa = 3.88ITTEE621 pKa = 4.03DD622 pKa = 3.47SAEE625 pKa = 3.9TFMANVPDD633 pKa = 5.17DD634 pKa = 3.75STIANTPQIQMTLGSTDD651 pKa = 3.63EE652 pKa = 4.59PSTDD656 pKa = 3.38TVHH659 pKa = 7.41SEE661 pKa = 4.03TTISNTLGLKK671 pKa = 8.92LTTEE675 pKa = 4.37GVTYY679 pKa = 11.02ASTTNVSDD687 pKa = 2.62SWTISNTPEE696 pKa = 3.78TKK698 pKa = 10.63LITEE702 pKa = 4.34STEE705 pKa = 3.88KK706 pKa = 10.76CCTSNVPDD714 pKa = 4.04YY715 pKa = 10.86STISDD720 pKa = 3.48TSGIEE725 pKa = 4.05LPTEE729 pKa = 4.15STTLASVNNGPYY741 pKa = 9.96HH742 pKa = 5.67STVSSTPHH750 pKa = 7.43AEE752 pKa = 3.78FTTEE756 pKa = 3.08ITAEE760 pKa = 4.09VSTTNAPSHH769 pKa = 5.37STVLNTLNVEE779 pKa = 4.3LTTEE783 pKa = 4.15GAVDD787 pKa = 3.51VSTANVPDD795 pKa = 5.43FSSIPYY801 pKa = 9.35TSDD804 pKa = 3.22VEE806 pKa = 4.44MNTDD810 pKa = 3.26NTVDD814 pKa = 3.06ASTTNRR820 pKa = 11.84PDD822 pKa = 2.74HH823 pKa = 5.33STIRR827 pKa = 11.84NIPASQSTTDD837 pKa = 3.2GSEE840 pKa = 4.29EE841 pKa = 4.02VSTTYY846 pKa = 11.15VLDD849 pKa = 3.87HH850 pKa = 6.51SVVPSMPEE858 pKa = 3.44IDD860 pKa = 3.44WTTKK864 pKa = 5.48TTEE867 pKa = 3.71GALPIRR873 pKa = 11.84FSAKK877 pKa = 8.44VTTISYY883 pKa = 6.1TTKK886 pKa = 10.81SPMATMEE893 pKa = 4.38STFSSLDD900 pKa = 3.53NNEE903 pKa = 3.91LTTTLVVTSSSTLSTDD919 pKa = 2.86SSFEE923 pKa = 4.06SSSDD927 pKa = 3.6GPSTSIEE934 pKa = 4.27TTTSFSLDD942 pKa = 3.42AVPEE946 pKa = 4.12ASASIAAIVSAVFGRR961 pKa = 11.84CVVCYY966 pKa = 9.72LVILCVNN973 pKa = 4.18
MM1 pKa = 7.31NSLFKK6 pKa = 10.45FCAIVTVLLDD16 pKa = 3.87LSTYY20 pKa = 9.74NPRR23 pKa = 11.84GSNTYY28 pKa = 8.31NTDD31 pKa = 3.75RR32 pKa = 11.84IISTKK37 pKa = 9.68IDD39 pKa = 3.31AAQSILTTNLSFHH52 pKa = 7.03LATEE56 pKa = 4.32TTTLFSTTVSNTITSSTDD74 pKa = 3.02SFPDD78 pKa = 2.79RR79 pKa = 11.84STIVNTPEE87 pKa = 3.58THH89 pKa = 5.89LTTGSSEE96 pKa = 4.13EE97 pKa = 4.2SSTLNIIDD105 pKa = 5.39GYY107 pKa = 8.07TISYY111 pKa = 10.38APDD114 pKa = 3.8AEE116 pKa = 4.44LTTEE120 pKa = 4.03ITAEE124 pKa = 4.15TSATNAPDD132 pKa = 3.27RR133 pKa = 11.84STIPNRR139 pKa = 11.84PNTQLTTDD147 pKa = 3.41TTEE150 pKa = 3.77QSFTTNIPYY159 pKa = 10.7DD160 pKa = 3.6LTIPLSKK167 pKa = 10.83VKK169 pKa = 9.54LTTEE173 pKa = 4.76GIEE176 pKa = 4.01EE177 pKa = 5.55GYY179 pKa = 6.78TTTGYY184 pKa = 8.56STIPDD189 pKa = 3.59TSGTEE194 pKa = 3.97LPTEE198 pKa = 4.28STTGAITNTASNHH211 pKa = 4.47STLLSNSEE219 pKa = 4.17VEE221 pKa = 4.44LTTEE225 pKa = 4.07STTDD229 pKa = 3.18ASITNLSDD237 pKa = 3.14NLTISNTVGAEE248 pKa = 3.83IITEE252 pKa = 4.3GTVEE256 pKa = 4.05TSTTNVPNDD265 pKa = 3.56SASPNVPEE273 pKa = 4.09TSITSGTTQHH283 pKa = 6.94PFLTSIPDD291 pKa = 3.79DD292 pKa = 3.71STVSYY297 pKa = 10.39TPGAEE302 pKa = 3.98LTTEE306 pKa = 4.26GTKK309 pKa = 9.51DD310 pKa = 2.93TSTINLSDD318 pKa = 3.8NLTISSTTKK327 pKa = 10.67AKK329 pKa = 10.3IITEE333 pKa = 4.3GTVEE337 pKa = 4.05TSTTNVPNDD346 pKa = 3.56SASPNVPEE354 pKa = 4.31TSTTSGTTQHH364 pKa = 6.97PFLTSIPDD372 pKa = 3.79DD373 pKa = 3.71STVSYY378 pKa = 10.39TPGAEE383 pKa = 4.02LTTEE387 pKa = 4.27STTDD391 pKa = 2.96TYY393 pKa = 9.04TTNLSDD399 pKa = 4.87NLTIPNTAGAEE410 pKa = 4.03ITTEE414 pKa = 4.29GSAEE418 pKa = 4.01TSTTNVPKK426 pKa = 10.76DD427 pKa = 3.43STIPNAPEE435 pKa = 3.46IQMTLASTDD444 pKa = 3.32VHH446 pKa = 6.34STDD449 pKa = 3.64TDD451 pKa = 3.5HH452 pKa = 7.17GNSTISYY459 pKa = 7.93TPGVEE464 pKa = 3.96LTTEE468 pKa = 3.62ITTDD472 pKa = 3.1AFSTHH477 pKa = 6.87PSDD480 pKa = 5.23HH481 pKa = 5.99LTIPNTIEE489 pKa = 4.37AEE491 pKa = 3.78ITTEE495 pKa = 4.01VNGDD499 pKa = 3.6STISNMPVTEE509 pKa = 4.37LTTEE513 pKa = 4.12STTDD517 pKa = 2.99ASTTNLSDD525 pKa = 3.53NLTIPNTTGAEE536 pKa = 3.89ITTEE540 pKa = 4.27GSAEE544 pKa = 4.03TSTTNIPNDD553 pKa = 3.38STIPNISDD561 pKa = 2.81IQMALGSTDD570 pKa = 3.66EE571 pKa = 4.65PSTDD575 pKa = 3.44TVHH578 pKa = 6.9GDD580 pKa = 3.2STISYY585 pKa = 8.67TPGAEE590 pKa = 4.0LTTEE594 pKa = 4.19STTDD598 pKa = 2.86TSTINLSDD606 pKa = 3.68NLIIPNTTGAEE617 pKa = 3.88ITTEE621 pKa = 4.03DD622 pKa = 3.47SAEE625 pKa = 3.9TFMANVPDD633 pKa = 5.17DD634 pKa = 3.75STIANTPQIQMTLGSTDD651 pKa = 3.63EE652 pKa = 4.59PSTDD656 pKa = 3.38TVHH659 pKa = 7.41SEE661 pKa = 4.03TTISNTLGLKK671 pKa = 8.92LTTEE675 pKa = 4.37GVTYY679 pKa = 11.02ASTTNVSDD687 pKa = 2.62SWTISNTPEE696 pKa = 3.78TKK698 pKa = 10.63LITEE702 pKa = 4.34STEE705 pKa = 3.88KK706 pKa = 10.76CCTSNVPDD714 pKa = 4.04YY715 pKa = 10.86STISDD720 pKa = 3.48TSGIEE725 pKa = 4.05LPTEE729 pKa = 4.15STTLASVNNGPYY741 pKa = 9.96HH742 pKa = 5.67STVSSTPHH750 pKa = 7.43AEE752 pKa = 3.78FTTEE756 pKa = 3.08ITAEE760 pKa = 4.09VSTTNAPSHH769 pKa = 5.37STVLNTLNVEE779 pKa = 4.3LTTEE783 pKa = 4.15GAVDD787 pKa = 3.51VSTANVPDD795 pKa = 5.43FSSIPYY801 pKa = 9.35TSDD804 pKa = 3.22VEE806 pKa = 4.44MNTDD810 pKa = 3.26NTVDD814 pKa = 3.06ASTTNRR820 pKa = 11.84PDD822 pKa = 2.74HH823 pKa = 5.33STIRR827 pKa = 11.84NIPASQSTTDD837 pKa = 3.2GSEE840 pKa = 4.29EE841 pKa = 4.02VSTTYY846 pKa = 11.15VLDD849 pKa = 3.87HH850 pKa = 6.51SVVPSMPEE858 pKa = 3.44IDD860 pKa = 3.44WTTKK864 pKa = 5.48TTEE867 pKa = 3.71GALPIRR873 pKa = 11.84FSAKK877 pKa = 8.44VTTISYY883 pKa = 6.1TTKK886 pKa = 10.81SPMATMEE893 pKa = 4.38STFSSLDD900 pKa = 3.53NNEE903 pKa = 3.91LTTTLVVTSSSTLSTDD919 pKa = 2.86SSFEE923 pKa = 4.06SSSDD927 pKa = 3.6GPSTSIEE934 pKa = 4.27TTTSFSLDD942 pKa = 3.42AVPEE946 pKa = 4.12ASASIAAIVSAVFGRR961 pKa = 11.84CVVCYY966 pKa = 9.72LVILCVNN973 pKa = 4.18
Molecular weight: 102.27 kDa
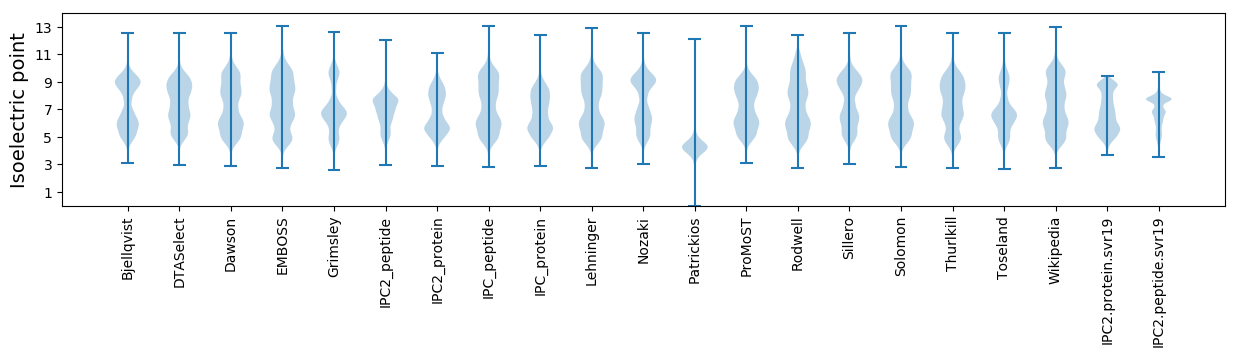
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W4X2L2|A0A1W4X2L2_AGRPL LOW QUALITY PROTEIN: serine palmitoyltransferase 1 OS=Agrilus planipennis OX=224129 GN=LOC108740496 PE=4 SV=2
MM1 pKa = 7.44SAHH4 pKa = 6.1KK5 pKa = 9.54TFIIKK10 pKa = 10.29RR11 pKa = 11.84KK12 pKa = 8.65LAKK15 pKa = 9.89KK16 pKa = 10.19LKK18 pKa = 8.64QNRR21 pKa = 11.84PIPQWVRR28 pKa = 11.84MRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.1WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.89LKK50 pKa = 10.55LL51 pKa = 3.39
MM1 pKa = 7.44SAHH4 pKa = 6.1KK5 pKa = 9.54TFIIKK10 pKa = 10.29RR11 pKa = 11.84KK12 pKa = 8.65LAKK15 pKa = 9.89KK16 pKa = 10.19LKK18 pKa = 8.64QNRR21 pKa = 11.84PIPQWVRR28 pKa = 11.84MRR30 pKa = 11.84TGNTIRR36 pKa = 11.84YY37 pKa = 5.79NAKK40 pKa = 8.47RR41 pKa = 11.84RR42 pKa = 11.84HH43 pKa = 4.1WRR45 pKa = 11.84RR46 pKa = 11.84TKK48 pKa = 10.89LKK50 pKa = 10.55LL51 pKa = 3.39
Molecular weight: 6.36 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
11197405 |
30 |
16983 |
596.5 |
67.29 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.483 ± 0.016 | 2.168 ± 0.063 |
5.302 ± 0.015 | 6.922 ± 0.028 |
3.885 ± 0.017 | 5.283 ± 0.021 |
2.364 ± 0.01 | 6.008 ± 0.015 |
7.099 ± 0.031 | 8.862 ± 0.036 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.012 | 5.692 ± 0.022 |
5.196 ± 0.036 | 4.335 ± 0.019 |
4.869 ± 0.016 | 8.303 ± 0.024 |
5.865 ± 0.015 | 6.066 ± 0.014 |
1.003 ± 0.008 | 3.217 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
