
Desulfosoma caldarium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; delta/epsilon subdivisions; Deltaproteobacteria; Syntrophobacterales; Syntrophobacteraceae; Desulfosoma
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins
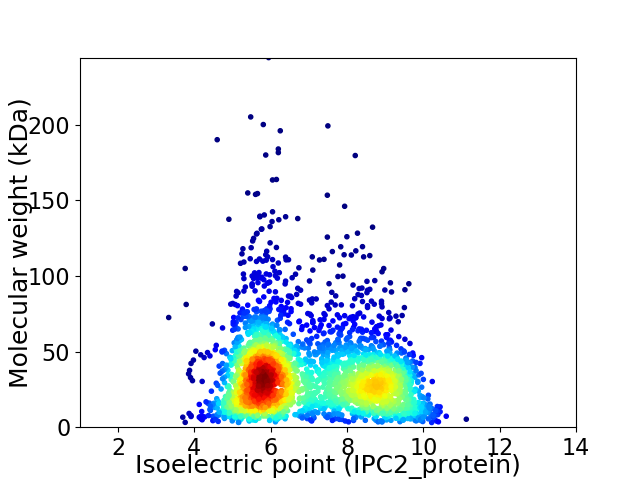
Virtual 2D-PAGE plot for 2924 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N1UQS2|A0A3N1UQS2_9DELT SNF2 domain-containing protein OS=Desulfosoma caldarium OX=610254 GN=EDC27_2161 PE=4 SV=1
MM1 pKa = 6.54VTALPPSSFLYY12 pKa = 9.59WAGPGLGYY20 pKa = 10.82DD21 pKa = 3.5GVVLVRR27 pKa = 11.84TDD29 pKa = 3.09DD30 pKa = 3.77GYY32 pKa = 11.49YY33 pKa = 10.48GSGVLLYY40 pKa = 10.87DD41 pKa = 4.09GGAVLTAAHH50 pKa = 6.47LVARR54 pKa = 11.84NGLPGAVVDD63 pKa = 3.78QAEE66 pKa = 4.67VVFQTTDD73 pKa = 2.85GEE75 pKa = 4.3YY76 pKa = 10.42SYY78 pKa = 10.94ISQEE82 pKa = 3.48ISVPTSYY89 pKa = 11.24DD90 pKa = 3.45PEE92 pKa = 4.28NLNGDD97 pKa = 4.39LALIWLPSTAPVSAEE112 pKa = 3.57RR113 pKa = 11.84YY114 pKa = 7.85EE115 pKa = 4.44LYY117 pKa = 10.26RR118 pKa = 11.84SHH120 pKa = 8.22DD121 pKa = 3.81EE122 pKa = 3.7IGRR125 pKa = 11.84VMTLVGYY132 pKa = 7.47GVPGDD137 pKa = 4.08GFYY140 pKa = 11.43GMYY143 pKa = 9.69EE144 pKa = 3.97NWSGPPLRR152 pKa = 11.84LRR154 pKa = 11.84AWNRR158 pKa = 11.84ADD160 pKa = 4.17ADD162 pKa = 3.65AGTVKK167 pKa = 10.6EE168 pKa = 4.0GLGYY172 pKa = 11.25VMDD175 pKa = 4.75WDD177 pKa = 4.12PKK179 pKa = 10.57EE180 pKa = 4.99DD181 pKa = 3.73SQLVADD187 pKa = 5.13FDD189 pKa = 5.04DD190 pKa = 3.83GTMVHH195 pKa = 7.04DD196 pKa = 5.15AMGSLLGVWDD206 pKa = 4.9LGLGYY211 pKa = 10.13YY212 pKa = 10.3EE213 pKa = 5.02GMISSGDD220 pKa = 3.42SGGPAFIEE228 pKa = 4.49GKK230 pKa = 9.69VAGIATAHH238 pKa = 6.76ASLGFWNIWPDD249 pKa = 2.95VDD251 pKa = 3.45IFKK254 pKa = 10.78NSSFGEE260 pKa = 3.81MGFWQRR266 pKa = 11.84VSSYY270 pKa = 9.56QEE272 pKa = 3.69WIDD275 pKa = 3.35RR276 pKa = 11.84SLRR279 pKa = 11.84SHH281 pKa = 6.52YY282 pKa = 10.19PNAPSRR288 pKa = 11.84PEE290 pKa = 4.24DD291 pKa = 3.39VQKK294 pKa = 10.87RR295 pKa = 11.84VTEE298 pKa = 4.27GDD300 pKa = 2.91AGTTYY305 pKa = 10.99AYY307 pKa = 10.72FLLQFNGVRR316 pKa = 11.84SDD318 pKa = 3.4PNAWVSVDD326 pKa = 3.12YY327 pKa = 7.47EE328 pKa = 4.5TRR330 pKa = 11.84DD331 pKa = 3.47GTATAGLDD339 pKa = 3.55YY340 pKa = 11.19VPASGTLILYY350 pKa = 9.36PGEE353 pKa = 4.82DD354 pKa = 3.7YY355 pKa = 11.44AVIPVEE361 pKa = 4.37IIGDD365 pKa = 3.97TVPEE369 pKa = 4.13PDD371 pKa = 3.07EE372 pKa = 4.51TFFLDD377 pKa = 3.08VFNPVGGSFGEE388 pKa = 4.75GVTKK392 pKa = 9.37LTAMRR397 pKa = 11.84TIVDD401 pKa = 3.91DD402 pKa = 5.79DD403 pKa = 4.39VSFWAA408 pKa = 4.83
MM1 pKa = 6.54VTALPPSSFLYY12 pKa = 9.59WAGPGLGYY20 pKa = 10.82DD21 pKa = 3.5GVVLVRR27 pKa = 11.84TDD29 pKa = 3.09DD30 pKa = 3.77GYY32 pKa = 11.49YY33 pKa = 10.48GSGVLLYY40 pKa = 10.87DD41 pKa = 4.09GGAVLTAAHH50 pKa = 6.47LVARR54 pKa = 11.84NGLPGAVVDD63 pKa = 3.78QAEE66 pKa = 4.67VVFQTTDD73 pKa = 2.85GEE75 pKa = 4.3YY76 pKa = 10.42SYY78 pKa = 10.94ISQEE82 pKa = 3.48ISVPTSYY89 pKa = 11.24DD90 pKa = 3.45PEE92 pKa = 4.28NLNGDD97 pKa = 4.39LALIWLPSTAPVSAEE112 pKa = 3.57RR113 pKa = 11.84YY114 pKa = 7.85EE115 pKa = 4.44LYY117 pKa = 10.26RR118 pKa = 11.84SHH120 pKa = 8.22DD121 pKa = 3.81EE122 pKa = 3.7IGRR125 pKa = 11.84VMTLVGYY132 pKa = 7.47GVPGDD137 pKa = 4.08GFYY140 pKa = 11.43GMYY143 pKa = 9.69EE144 pKa = 3.97NWSGPPLRR152 pKa = 11.84LRR154 pKa = 11.84AWNRR158 pKa = 11.84ADD160 pKa = 4.17ADD162 pKa = 3.65AGTVKK167 pKa = 10.6EE168 pKa = 4.0GLGYY172 pKa = 11.25VMDD175 pKa = 4.75WDD177 pKa = 4.12PKK179 pKa = 10.57EE180 pKa = 4.99DD181 pKa = 3.73SQLVADD187 pKa = 5.13FDD189 pKa = 5.04DD190 pKa = 3.83GTMVHH195 pKa = 7.04DD196 pKa = 5.15AMGSLLGVWDD206 pKa = 4.9LGLGYY211 pKa = 10.13YY212 pKa = 10.3EE213 pKa = 5.02GMISSGDD220 pKa = 3.42SGGPAFIEE228 pKa = 4.49GKK230 pKa = 9.69VAGIATAHH238 pKa = 6.76ASLGFWNIWPDD249 pKa = 2.95VDD251 pKa = 3.45IFKK254 pKa = 10.78NSSFGEE260 pKa = 3.81MGFWQRR266 pKa = 11.84VSSYY270 pKa = 9.56QEE272 pKa = 3.69WIDD275 pKa = 3.35RR276 pKa = 11.84SLRR279 pKa = 11.84SHH281 pKa = 6.52YY282 pKa = 10.19PNAPSRR288 pKa = 11.84PEE290 pKa = 4.24DD291 pKa = 3.39VQKK294 pKa = 10.87RR295 pKa = 11.84VTEE298 pKa = 4.27GDD300 pKa = 2.91AGTTYY305 pKa = 10.99AYY307 pKa = 10.72FLLQFNGVRR316 pKa = 11.84SDD318 pKa = 3.4PNAWVSVDD326 pKa = 3.12YY327 pKa = 7.47EE328 pKa = 4.5TRR330 pKa = 11.84DD331 pKa = 3.47GTATAGLDD339 pKa = 3.55YY340 pKa = 11.19VPASGTLILYY350 pKa = 9.36PGEE353 pKa = 4.82DD354 pKa = 3.7YY355 pKa = 11.44AVIPVEE361 pKa = 4.37IIGDD365 pKa = 3.97TVPEE369 pKa = 4.13PDD371 pKa = 3.07EE372 pKa = 4.51TFFLDD377 pKa = 3.08VFNPVGGSFGEE388 pKa = 4.75GVTKK392 pKa = 9.37LTAMRR397 pKa = 11.84TIVDD401 pKa = 3.91DD402 pKa = 5.79DD403 pKa = 4.39VSFWAA408 pKa = 4.83
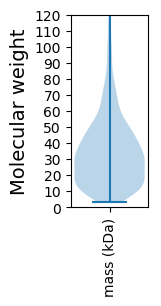
Molecular weight: 44.37 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N1VLD6|A0A3N1VLD6_9DELT Metallo-beta-lactamase family protein OS=Desulfosoma caldarium OX=610254 GN=EDC27_0957 PE=4 SV=1
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.03QPHH8 pKa = 5.72NLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.76RR14 pKa = 11.84THH16 pKa = 5.88GFLVRR21 pKa = 11.84MATKK25 pKa = 10.34SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.4GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
MM1 pKa = 7.28KK2 pKa = 9.44RR3 pKa = 11.84TYY5 pKa = 10.03QPHH8 pKa = 5.72NLRR11 pKa = 11.84RR12 pKa = 11.84KK13 pKa = 7.76RR14 pKa = 11.84THH16 pKa = 5.88GFLVRR21 pKa = 11.84MATKK25 pKa = 10.34SGRR28 pKa = 11.84AVINRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.4GRR39 pKa = 11.84KK40 pKa = 8.55RR41 pKa = 11.84LSVV44 pKa = 3.2
Molecular weight: 5.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
962980 |
29 |
2176 |
329.3 |
36.59 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.713 ± 0.052 | 1.367 ± 0.022 |
5.058 ± 0.038 | 6.537 ± 0.05 |
3.981 ± 0.032 | 7.57 ± 0.042 |
2.57 ± 0.02 | 4.8 ± 0.037 |
4.294 ± 0.036 | 10.825 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.548 ± 0.02 | 2.461 ± 0.022 |
5.111 ± 0.029 | 3.575 ± 0.026 |
7.338 ± 0.042 | 5.161 ± 0.03 |
4.839 ± 0.024 | 8.243 ± 0.04 |
1.409 ± 0.019 | 2.599 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
