
Microbacterium aerolatum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Microbacteriaceae; Microbacterium
Average proteome isoelectric point is 5.9
Get precalculated fractions of proteins
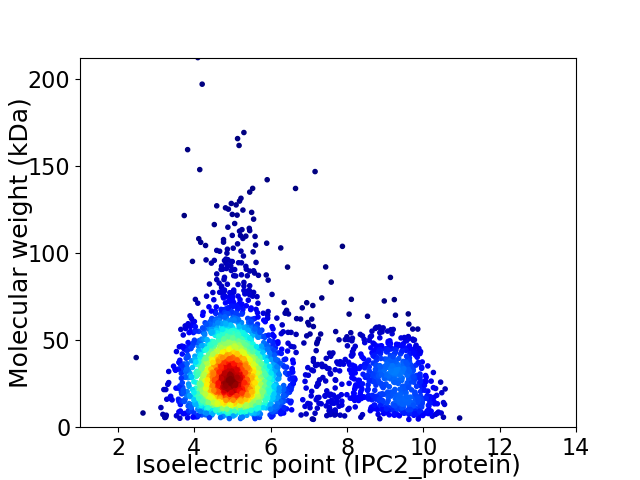
Virtual 2D-PAGE plot for 3137 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A511AC89|A0A511AC89_9MICO DNA-binding response regulator OS=Microbacterium aerolatum OX=153731 GN=MAE01_09510 PE=4 SV=1
MM1 pKa = 7.36HH2 pKa = 7.14SKK4 pKa = 10.63ARR6 pKa = 11.84LVAVGSAVAALALAGCSAGQANDD29 pKa = 3.79GGGDD33 pKa = 3.39SADD36 pKa = 4.13PIRR39 pKa = 11.84YY40 pKa = 8.86AVEE43 pKa = 4.25QPDD46 pKa = 3.76SMIPGNQFSSYY57 pKa = 10.83VILEE61 pKa = 4.5TIFAPLTQVDD71 pKa = 3.78EE72 pKa = 4.91DD73 pKa = 4.91GEE75 pKa = 4.7VSFVAAEE82 pKa = 4.22SVEE85 pKa = 4.36SDD87 pKa = 4.69DD88 pKa = 3.98AQTWTITLRR97 pKa = 11.84DD98 pKa = 3.26GWTFHH103 pKa = 7.39DD104 pKa = 4.96GEE106 pKa = 4.71PVTAQDD112 pKa = 6.11YY113 pKa = 10.51VDD115 pKa = 3.3TWNYY119 pKa = 9.32AAYY122 pKa = 9.96GPNAWVNGPQLANVAGYY139 pKa = 11.32ADD141 pKa = 4.85LNPAEE146 pKa = 4.78GEE148 pKa = 4.08PTATEE153 pKa = 3.88LAGLSVVDD161 pKa = 4.77DD162 pKa = 3.56RR163 pKa = 11.84TFTVEE168 pKa = 4.47LSAPDD173 pKa = 3.39RR174 pKa = 11.84QYY176 pKa = 11.59PLQLGIGQTGLYY188 pKa = 8.18PLPSEE193 pKa = 5.16ALDD196 pKa = 5.46DD197 pKa = 3.93LAAWEE202 pKa = 4.29EE203 pKa = 4.31APVGNGPFEE212 pKa = 4.4LTDD215 pKa = 3.33GWSEE219 pKa = 4.06SDD221 pKa = 3.93PIEE224 pKa = 4.06AVAYY228 pKa = 10.28DD229 pKa = 5.11DD230 pKa = 4.26YY231 pKa = 12.04AGDD234 pKa = 4.48APTLDD239 pKa = 3.21AVTFVPYY246 pKa = 10.77LDD248 pKa = 3.6TATAYY253 pKa = 9.58TDD255 pKa = 4.08ALAGDD260 pKa = 3.92IDD262 pKa = 3.8IAAIAAGQAVQARR275 pKa = 11.84NDD277 pKa = 3.65FGDD280 pKa = 3.51SVYY283 pKa = 10.98EE284 pKa = 3.84LDD286 pKa = 4.26APGVDD291 pKa = 4.12FLGFTLSDD299 pKa = 3.31PRR301 pKa = 11.84FSDD304 pKa = 3.12VRR306 pKa = 11.84VRR308 pKa = 11.84QAISMAIDD316 pKa = 3.26RR317 pKa = 11.84EE318 pKa = 4.48AINDD322 pKa = 3.71AVFGGSQVPATSLTAPSMPGDD343 pKa = 3.67PEE345 pKa = 4.75GVCGEE350 pKa = 4.2YY351 pKa = 10.99CEE353 pKa = 5.03FDD355 pKa = 3.66PDD357 pKa = 3.47AAKK360 pKa = 10.85ALLEE364 pKa = 4.11EE365 pKa = 4.8AGGFDD370 pKa = 4.8GEE372 pKa = 4.57MKK374 pKa = 10.5VHH376 pKa = 6.9FVANWGQEE384 pKa = 4.0DD385 pKa = 4.59YY386 pKa = 11.43FEE388 pKa = 6.6AIANQLRR395 pKa = 11.84QNLGIEE401 pKa = 4.28AVIASPSADD410 pKa = 2.97FAAFTDD416 pKa = 4.17AVQNGEE422 pKa = 3.75TDD424 pKa = 3.61GPFRR428 pKa = 11.84ARR430 pKa = 11.84WGALYY435 pKa = 9.57PSQQNTLTAVFTATGEE451 pKa = 4.54GNFGAGGYY459 pKa = 9.65SSPEE463 pKa = 3.5VDD465 pKa = 3.3EE466 pKa = 5.56LLRR469 pKa = 11.84QANAADD475 pKa = 3.77TLEE478 pKa = 4.89DD479 pKa = 3.57SLAGYY484 pKa = 9.31VAAQEE489 pKa = 5.37RR490 pKa = 11.84ILQDD494 pKa = 3.71FPTVPMFGNRR504 pKa = 11.84YY505 pKa = 9.82LYY507 pKa = 9.1VTSDD511 pKa = 4.41RR512 pKa = 11.84IAEE515 pKa = 4.06LHH517 pKa = 6.06TNAGAIDD524 pKa = 3.78VPKK527 pKa = 10.39IAVAGG532 pKa = 3.88
MM1 pKa = 7.36HH2 pKa = 7.14SKK4 pKa = 10.63ARR6 pKa = 11.84LVAVGSAVAALALAGCSAGQANDD29 pKa = 3.79GGGDD33 pKa = 3.39SADD36 pKa = 4.13PIRR39 pKa = 11.84YY40 pKa = 8.86AVEE43 pKa = 4.25QPDD46 pKa = 3.76SMIPGNQFSSYY57 pKa = 10.83VILEE61 pKa = 4.5TIFAPLTQVDD71 pKa = 3.78EE72 pKa = 4.91DD73 pKa = 4.91GEE75 pKa = 4.7VSFVAAEE82 pKa = 4.22SVEE85 pKa = 4.36SDD87 pKa = 4.69DD88 pKa = 3.98AQTWTITLRR97 pKa = 11.84DD98 pKa = 3.26GWTFHH103 pKa = 7.39DD104 pKa = 4.96GEE106 pKa = 4.71PVTAQDD112 pKa = 6.11YY113 pKa = 10.51VDD115 pKa = 3.3TWNYY119 pKa = 9.32AAYY122 pKa = 9.96GPNAWVNGPQLANVAGYY139 pKa = 11.32ADD141 pKa = 4.85LNPAEE146 pKa = 4.78GEE148 pKa = 4.08PTATEE153 pKa = 3.88LAGLSVVDD161 pKa = 4.77DD162 pKa = 3.56RR163 pKa = 11.84TFTVEE168 pKa = 4.47LSAPDD173 pKa = 3.39RR174 pKa = 11.84QYY176 pKa = 11.59PLQLGIGQTGLYY188 pKa = 8.18PLPSEE193 pKa = 5.16ALDD196 pKa = 5.46DD197 pKa = 3.93LAAWEE202 pKa = 4.29EE203 pKa = 4.31APVGNGPFEE212 pKa = 4.4LTDD215 pKa = 3.33GWSEE219 pKa = 4.06SDD221 pKa = 3.93PIEE224 pKa = 4.06AVAYY228 pKa = 10.28DD229 pKa = 5.11DD230 pKa = 4.26YY231 pKa = 12.04AGDD234 pKa = 4.48APTLDD239 pKa = 3.21AVTFVPYY246 pKa = 10.77LDD248 pKa = 3.6TATAYY253 pKa = 9.58TDD255 pKa = 4.08ALAGDD260 pKa = 3.92IDD262 pKa = 3.8IAAIAAGQAVQARR275 pKa = 11.84NDD277 pKa = 3.65FGDD280 pKa = 3.51SVYY283 pKa = 10.98EE284 pKa = 3.84LDD286 pKa = 4.26APGVDD291 pKa = 4.12FLGFTLSDD299 pKa = 3.31PRR301 pKa = 11.84FSDD304 pKa = 3.12VRR306 pKa = 11.84VRR308 pKa = 11.84QAISMAIDD316 pKa = 3.26RR317 pKa = 11.84EE318 pKa = 4.48AINDD322 pKa = 3.71AVFGGSQVPATSLTAPSMPGDD343 pKa = 3.67PEE345 pKa = 4.75GVCGEE350 pKa = 4.2YY351 pKa = 10.99CEE353 pKa = 5.03FDD355 pKa = 3.66PDD357 pKa = 3.47AAKK360 pKa = 10.85ALLEE364 pKa = 4.11EE365 pKa = 4.8AGGFDD370 pKa = 4.8GEE372 pKa = 4.57MKK374 pKa = 10.5VHH376 pKa = 6.9FVANWGQEE384 pKa = 4.0DD385 pKa = 4.59YY386 pKa = 11.43FEE388 pKa = 6.6AIANQLRR395 pKa = 11.84QNLGIEE401 pKa = 4.28AVIASPSADD410 pKa = 2.97FAAFTDD416 pKa = 4.17AVQNGEE422 pKa = 3.75TDD424 pKa = 3.61GPFRR428 pKa = 11.84ARR430 pKa = 11.84WGALYY435 pKa = 9.57PSQQNTLTAVFTATGEE451 pKa = 4.54GNFGAGGYY459 pKa = 9.65SSPEE463 pKa = 3.5VDD465 pKa = 3.3EE466 pKa = 5.56LLRR469 pKa = 11.84QANAADD475 pKa = 3.77TLEE478 pKa = 4.89DD479 pKa = 3.57SLAGYY484 pKa = 9.31VAAQEE489 pKa = 5.37RR490 pKa = 11.84ILQDD494 pKa = 3.71FPTVPMFGNRR504 pKa = 11.84YY505 pKa = 9.82LYY507 pKa = 9.1VTSDD511 pKa = 4.41RR512 pKa = 11.84IAEE515 pKa = 4.06LHH517 pKa = 6.06TNAGAIDD524 pKa = 3.78VPKK527 pKa = 10.39IAVAGG532 pKa = 3.88
Molecular weight: 56.21 kDa
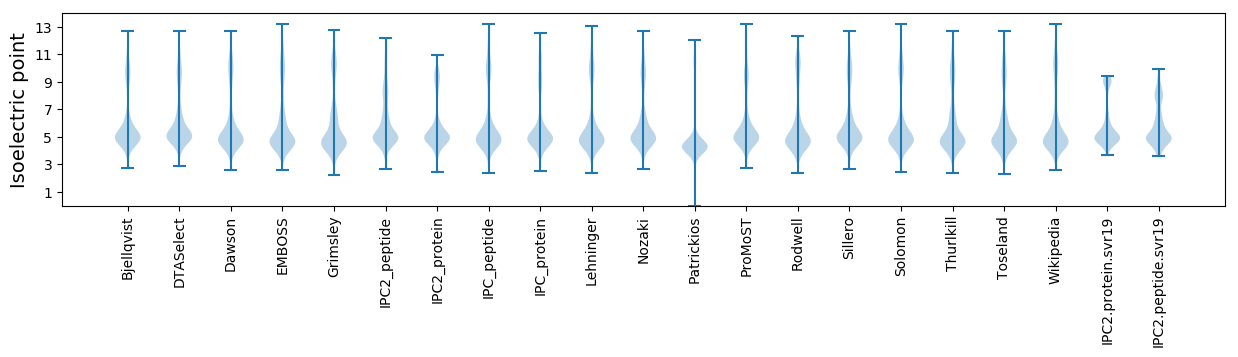
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A511AEF6|A0A511AEF6_9MICO DNA helicase OS=Microbacterium aerolatum OX=153731 GN=MAE01_17240 PE=4 SV=1
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
MM1 pKa = 7.69SKK3 pKa = 9.0RR4 pKa = 11.84TFQPNNRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84AKK15 pKa = 10.04KK16 pKa = 8.86HH17 pKa = 4.36GFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TRR26 pKa = 11.84AGRR29 pKa = 11.84SILSARR35 pKa = 11.84RR36 pKa = 11.84AKK38 pKa = 10.56GRR40 pKa = 11.84TEE42 pKa = 4.14LSAA45 pKa = 4.86
Molecular weight: 5.27 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
990643 |
41 |
2029 |
315.8 |
33.89 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.305 ± 0.064 | 0.477 ± 0.009 |
6.409 ± 0.038 | 5.95 ± 0.035 |
3.152 ± 0.027 | 8.755 ± 0.036 |
2.009 ± 0.022 | 4.977 ± 0.035 |
2.07 ± 0.034 | 9.968 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.974 ± 0.018 | 2.004 ± 0.025 |
5.194 ± 0.03 | 2.871 ± 0.021 |
7.169 ± 0.054 | 5.606 ± 0.027 |
5.999 ± 0.031 | 8.666 ± 0.038 |
1.511 ± 0.018 | 1.934 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
