
Toxoplasma gondii (strain ATCC 50861 / VEG)
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Conoidasida; Coccidia; Eucoccidiorida; Eimeriorina; Sarcocystidae; Toxoplasma; Toxoplasma gondii; Toxoplasma gondii type III
Average proteome isoelectric point is 6.91
Get precalculated fractions of proteins
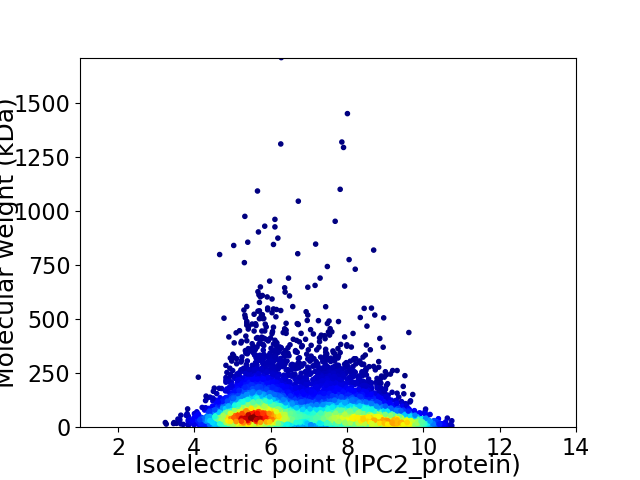
Virtual 2D-PAGE plot for 8404 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|V4YK67|V4YK67_TOXGV Uncharacterized protein (Fragment) OS=Toxoplasma gondii (strain ATCC 50861 / VEG) OX=432359 GN=TGVEG_361850 PE=4 SV=1
MM1 pKa = 7.02SAEE4 pKa = 4.27RR5 pKa = 11.84QSSSCDD11 pKa = 3.32CVLEE15 pKa = 4.32ATDD18 pKa = 4.28CASGTGNEE26 pKa = 4.14GVAFAVDD33 pKa = 3.72SFRR36 pKa = 11.84GFLPLEE42 pKa = 4.48VVCDD46 pKa = 4.35CRR48 pKa = 11.84PSPCDD53 pKa = 3.4GTLEE57 pKa = 4.42PADD60 pKa = 4.41PMDD63 pKa = 5.66SPGTWLVNDD72 pKa = 4.39FDD74 pKa = 4.99VDD76 pKa = 3.91FLRR79 pKa = 11.84ARR81 pKa = 11.84FPLEE85 pKa = 3.56EE86 pKa = 4.03TSAEE90 pKa = 4.11RR91 pKa = 11.84QSSSCDD97 pKa = 3.32CVLEE101 pKa = 4.32ATDD104 pKa = 4.28CASGTGNEE112 pKa = 4.14GVAFAVDD119 pKa = 3.72SFRR122 pKa = 11.84GFLPLEE128 pKa = 4.48VVCDD132 pKa = 4.35CRR134 pKa = 11.84PSPCDD139 pKa = 3.4GTLEE143 pKa = 4.42PADD146 pKa = 4.41PMDD149 pKa = 5.66SPGTWLVDD157 pKa = 3.68DD158 pKa = 5.07FDD160 pKa = 5.08VDD162 pKa = 3.88FLRR165 pKa = 11.84ARR167 pKa = 11.84FPLEE171 pKa = 3.56EE172 pKa = 4.03TSAEE176 pKa = 4.11RR177 pKa = 11.84QSSSCDD183 pKa = 3.32CVLEE187 pKa = 4.32ATDD190 pKa = 4.28CASGTGNEE198 pKa = 4.14GVAFAVDD205 pKa = 3.72SFRR208 pKa = 11.84GFLPLEE214 pKa = 4.48VVCDD218 pKa = 4.35CRR220 pKa = 11.84PSPCDD225 pKa = 3.4GTLEE229 pKa = 4.42PADD232 pKa = 4.41PMDD235 pKa = 5.66SPGTWLVDD243 pKa = 3.68DD244 pKa = 5.07FDD246 pKa = 5.08VDD248 pKa = 3.88FLRR251 pKa = 11.84ARR253 pKa = 11.84FPLGEE258 pKa = 4.1MSAEE262 pKa = 4.08RR263 pKa = 11.84QSSSCDD269 pKa = 3.32CVLEE273 pKa = 4.32ATDD276 pKa = 4.28CASGTGNEE284 pKa = 4.14GVAFAVDD291 pKa = 3.72SFRR294 pKa = 11.84GFLPLEE300 pKa = 4.48VVCDD304 pKa = 4.35CRR306 pKa = 11.84PSPCDD311 pKa = 3.4GTLEE315 pKa = 4.42PADD318 pKa = 4.57PMDD321 pKa = 4.16SPP323 pKa = 4.78
MM1 pKa = 7.02SAEE4 pKa = 4.27RR5 pKa = 11.84QSSSCDD11 pKa = 3.32CVLEE15 pKa = 4.32ATDD18 pKa = 4.28CASGTGNEE26 pKa = 4.14GVAFAVDD33 pKa = 3.72SFRR36 pKa = 11.84GFLPLEE42 pKa = 4.48VVCDD46 pKa = 4.35CRR48 pKa = 11.84PSPCDD53 pKa = 3.4GTLEE57 pKa = 4.42PADD60 pKa = 4.41PMDD63 pKa = 5.66SPGTWLVNDD72 pKa = 4.39FDD74 pKa = 4.99VDD76 pKa = 3.91FLRR79 pKa = 11.84ARR81 pKa = 11.84FPLEE85 pKa = 3.56EE86 pKa = 4.03TSAEE90 pKa = 4.11RR91 pKa = 11.84QSSSCDD97 pKa = 3.32CVLEE101 pKa = 4.32ATDD104 pKa = 4.28CASGTGNEE112 pKa = 4.14GVAFAVDD119 pKa = 3.72SFRR122 pKa = 11.84GFLPLEE128 pKa = 4.48VVCDD132 pKa = 4.35CRR134 pKa = 11.84PSPCDD139 pKa = 3.4GTLEE143 pKa = 4.42PADD146 pKa = 4.41PMDD149 pKa = 5.66SPGTWLVDD157 pKa = 3.68DD158 pKa = 5.07FDD160 pKa = 5.08VDD162 pKa = 3.88FLRR165 pKa = 11.84ARR167 pKa = 11.84FPLEE171 pKa = 3.56EE172 pKa = 4.03TSAEE176 pKa = 4.11RR177 pKa = 11.84QSSSCDD183 pKa = 3.32CVLEE187 pKa = 4.32ATDD190 pKa = 4.28CASGTGNEE198 pKa = 4.14GVAFAVDD205 pKa = 3.72SFRR208 pKa = 11.84GFLPLEE214 pKa = 4.48VVCDD218 pKa = 4.35CRR220 pKa = 11.84PSPCDD225 pKa = 3.4GTLEE229 pKa = 4.42PADD232 pKa = 4.41PMDD235 pKa = 5.66SPGTWLVDD243 pKa = 3.68DD244 pKa = 5.07FDD246 pKa = 5.08VDD248 pKa = 3.88FLRR251 pKa = 11.84ARR253 pKa = 11.84FPLGEE258 pKa = 4.1MSAEE262 pKa = 4.08RR263 pKa = 11.84QSSSCDD269 pKa = 3.32CVLEE273 pKa = 4.32ATDD276 pKa = 4.28CASGTGNEE284 pKa = 4.14GVAFAVDD291 pKa = 3.72SFRR294 pKa = 11.84GFLPLEE300 pKa = 4.48VVCDD304 pKa = 4.35CRR306 pKa = 11.84PSPCDD311 pKa = 3.4GTLEE315 pKa = 4.42PADD318 pKa = 4.57PMDD321 pKa = 4.16SPP323 pKa = 4.78
Molecular weight: 34.36 kDa
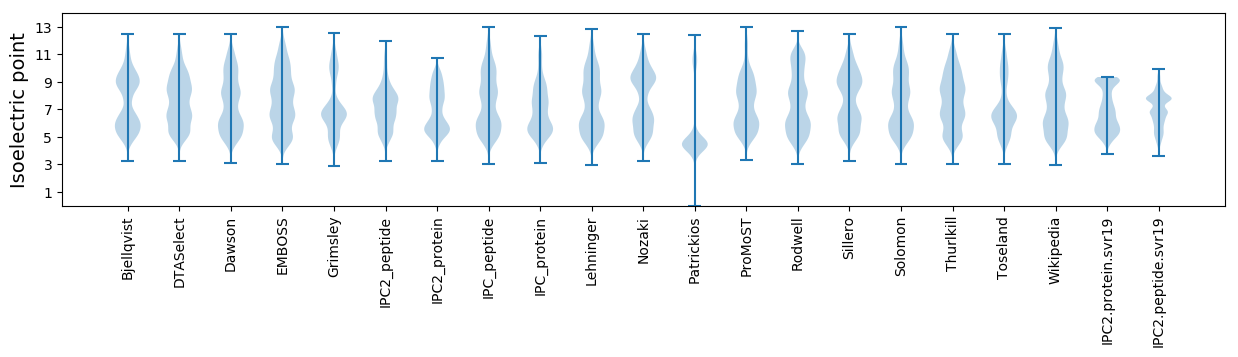
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A125YSL9|A0A125YSL9_TOXGV Uncharacterized protein OS=Toxoplasma gondii (strain ATCC 50861 / VEG) OX=432359 GN=BN1205_038190 PE=4 SV=1
MM1 pKa = 7.65LKK3 pKa = 10.06HH4 pKa = 6.47GSPIAQPRR12 pKa = 11.84RR13 pKa = 11.84SSRR16 pKa = 11.84VRR18 pKa = 11.84LPRR21 pKa = 11.84YY22 pKa = 8.1RR23 pKa = 11.84ARR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 10.85KK28 pKa = 9.75MDD30 pKa = 3.32STRR33 pKa = 11.84SSMSNEE39 pKa = 3.79AGLGQAVSHH48 pKa = 6.04QSTVSSSKK56 pKa = 10.2NPVFGGEE63 pKa = 4.3FFCLANPSATAAVFKK78 pKa = 10.71ASRR81 pKa = 11.84CARR84 pKa = 11.84CYY86 pKa = 10.62GPRR89 pKa = 11.84LSEE92 pKa = 3.69QGPFPRR98 pKa = 11.84EE99 pKa = 3.45EE100 pKa = 4.03ARR102 pKa = 11.84SVRR105 pKa = 11.84HH106 pKa = 5.22VRR108 pKa = 11.84PPGNPCGADD117 pKa = 3.26SQPRR121 pKa = 11.84CRR123 pKa = 11.84LPRR126 pKa = 11.84RR127 pKa = 11.84EE128 pKa = 4.11APRR131 pKa = 11.84GSRR134 pKa = 11.84GDD136 pKa = 3.55PGSPSGGQKK145 pKa = 9.76SAPGKK150 pKa = 9.23NVRR153 pKa = 11.84SNSDD157 pKa = 2.76SWPDD161 pKa = 3.28SRR163 pKa = 11.84NRR165 pKa = 11.84LPDD168 pKa = 3.59SQPTSASEE176 pKa = 4.2RR177 pKa = 11.84DD178 pKa = 3.68TGSPPLYY185 pKa = 10.57GCLCPRR191 pKa = 11.84SKK193 pKa = 10.9RR194 pKa = 11.84LGRR197 pKa = 11.84ASGRR201 pKa = 11.84AHH203 pKa = 7.12FYY205 pKa = 9.56ASEE208 pKa = 3.78WRR210 pKa = 11.84GGRR213 pKa = 11.84RR214 pKa = 11.84VFRR217 pKa = 11.84ASIEE221 pKa = 4.08PVGALGNKK229 pKa = 9.6SSATPRR235 pKa = 11.84KK236 pKa = 9.16GIRR239 pKa = 11.84VCGMEE244 pKa = 4.4LRR246 pKa = 11.84LNTPHH251 pKa = 7.09CPPPGCALLLGVLLLAASLLAVLFPSDD278 pKa = 3.39QVSRR282 pKa = 11.84HH283 pKa = 4.92IGKK286 pKa = 10.19FSFLNAISATEE297 pKa = 4.1LPQCLKK303 pKa = 10.83LAFSTSLGPHH313 pKa = 5.86PAWRR317 pKa = 11.84AVASAVFPLCGLAAGVAVVFFLFRR341 pKa = 11.84RR342 pKa = 11.84PYY344 pKa = 9.89IVEE347 pKa = 3.7QSLEE351 pKa = 3.75IVANFGVQLQQKK363 pKa = 6.31TRR365 pKa = 11.84WGGIKK370 pKa = 10.04RR371 pKa = 11.84QFIDD375 pKa = 3.41ASQVSDD381 pKa = 3.63VIINEE386 pKa = 4.03GFRR389 pKa = 11.84ACDD392 pKa = 3.23VVFYY396 pKa = 10.98VALLVKK402 pKa = 9.56DD403 pKa = 3.92QPSMVVPFQHH413 pKa = 6.67FPLPLEE419 pKa = 4.43EE420 pKa = 4.42NLVIYY425 pKa = 7.59EE426 pKa = 4.3TLHH429 pKa = 6.14HH430 pKa = 6.85LLHH433 pKa = 6.74ISSGVRR439 pKa = 11.84RR440 pKa = 11.84AA441 pKa = 3.2
MM1 pKa = 7.65LKK3 pKa = 10.06HH4 pKa = 6.47GSPIAQPRR12 pKa = 11.84RR13 pKa = 11.84SSRR16 pKa = 11.84VRR18 pKa = 11.84LPRR21 pKa = 11.84YY22 pKa = 8.1RR23 pKa = 11.84ARR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 10.85KK28 pKa = 9.75MDD30 pKa = 3.32STRR33 pKa = 11.84SSMSNEE39 pKa = 3.79AGLGQAVSHH48 pKa = 6.04QSTVSSSKK56 pKa = 10.2NPVFGGEE63 pKa = 4.3FFCLANPSATAAVFKK78 pKa = 10.71ASRR81 pKa = 11.84CARR84 pKa = 11.84CYY86 pKa = 10.62GPRR89 pKa = 11.84LSEE92 pKa = 3.69QGPFPRR98 pKa = 11.84EE99 pKa = 3.45EE100 pKa = 4.03ARR102 pKa = 11.84SVRR105 pKa = 11.84HH106 pKa = 5.22VRR108 pKa = 11.84PPGNPCGADD117 pKa = 3.26SQPRR121 pKa = 11.84CRR123 pKa = 11.84LPRR126 pKa = 11.84RR127 pKa = 11.84EE128 pKa = 4.11APRR131 pKa = 11.84GSRR134 pKa = 11.84GDD136 pKa = 3.55PGSPSGGQKK145 pKa = 9.76SAPGKK150 pKa = 9.23NVRR153 pKa = 11.84SNSDD157 pKa = 2.76SWPDD161 pKa = 3.28SRR163 pKa = 11.84NRR165 pKa = 11.84LPDD168 pKa = 3.59SQPTSASEE176 pKa = 4.2RR177 pKa = 11.84DD178 pKa = 3.68TGSPPLYY185 pKa = 10.57GCLCPRR191 pKa = 11.84SKK193 pKa = 10.9RR194 pKa = 11.84LGRR197 pKa = 11.84ASGRR201 pKa = 11.84AHH203 pKa = 7.12FYY205 pKa = 9.56ASEE208 pKa = 3.78WRR210 pKa = 11.84GGRR213 pKa = 11.84RR214 pKa = 11.84VFRR217 pKa = 11.84ASIEE221 pKa = 4.08PVGALGNKK229 pKa = 9.6SSATPRR235 pKa = 11.84KK236 pKa = 9.16GIRR239 pKa = 11.84VCGMEE244 pKa = 4.4LRR246 pKa = 11.84LNTPHH251 pKa = 7.09CPPPGCALLLGVLLLAASLLAVLFPSDD278 pKa = 3.39QVSRR282 pKa = 11.84HH283 pKa = 4.92IGKK286 pKa = 10.19FSFLNAISATEE297 pKa = 4.1LPQCLKK303 pKa = 10.83LAFSTSLGPHH313 pKa = 5.86PAWRR317 pKa = 11.84AVASAVFPLCGLAAGVAVVFFLFRR341 pKa = 11.84RR342 pKa = 11.84PYY344 pKa = 9.89IVEE347 pKa = 3.7QSLEE351 pKa = 3.75IVANFGVQLQQKK363 pKa = 6.31TRR365 pKa = 11.84WGGIKK370 pKa = 10.04RR371 pKa = 11.84QFIDD375 pKa = 3.41ASQVSDD381 pKa = 3.63VIINEE386 pKa = 4.03GFRR389 pKa = 11.84ACDD392 pKa = 3.23VVFYY396 pKa = 10.98VALLVKK402 pKa = 9.56DD403 pKa = 3.92QPSMVVPFQHH413 pKa = 6.67FPLPLEE419 pKa = 4.43EE420 pKa = 4.42NLVIYY425 pKa = 7.59EE426 pKa = 4.3TLHH429 pKa = 6.14HH430 pKa = 6.85LLHH433 pKa = 6.74ISSGVRR439 pKa = 11.84RR440 pKa = 11.84AA441 pKa = 3.2
Molecular weight: 48.11 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6625207 |
26 |
15897 |
788.3 |
85.67 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.209 ± 0.029 | 2.032 ± 0.014 |
4.726 ± 0.013 | 7.709 ± 0.034 |
3.475 ± 0.012 | 7.441 ± 0.032 |
2.181 ± 0.009 | 2.373 ± 0.015 |
4.018 ± 0.024 | 9.006 ± 0.028 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.4 ± 0.009 | 2.322 ± 0.013 |
6.464 ± 0.024 | 3.958 ± 0.015 |
8.265 ± 0.033 | 11.085 ± 0.041 |
5.042 ± 0.015 | 5.831 ± 0.018 |
1.014 ± 0.006 | 1.449 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
