
Candidatus Accumulibacter sp. SK-11
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Betaproteobacteria; Betaproteobacteria incertae sedis; Candidatus Accumulibacter; unclassified Candidatus Accumulibacter
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
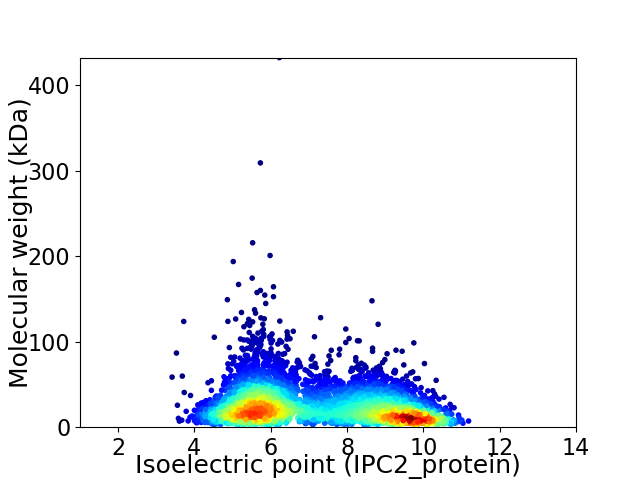
Virtual 2D-PAGE plot for 4767 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A011NK49|A0A011NK49_9PROT Arsenate reductase OS=Candidatus Accumulibacter sp. SK-11 OX=1454000 GN=arsC PE=3 SV=1
MM1 pKa = 7.78ASFSPNMPTTANGRR15 pKa = 11.84TVTSQGVYY23 pKa = 10.28SDD25 pKa = 4.81PLLPSYY31 pKa = 9.91WYY33 pKa = 10.92LGDD36 pKa = 3.56ASGAVKK42 pKa = 10.28GVNAMRR48 pKa = 11.84AWDD51 pKa = 4.24DD52 pKa = 3.49YY53 pKa = 10.08RR54 pKa = 11.84GSGIVVAVIDD64 pKa = 4.51DD65 pKa = 4.03GVEE68 pKa = 4.03YY69 pKa = 9.66THH71 pKa = 7.43LDD73 pKa = 2.93LAANYY78 pKa = 10.01RR79 pKa = 11.84SDD81 pKa = 3.38LAYY84 pKa = 8.8DD85 pKa = 3.53TRR87 pKa = 11.84DD88 pKa = 3.21RR89 pKa = 11.84DD90 pKa = 3.33ADD92 pKa = 3.75AFPGEE97 pKa = 4.17SSDD100 pKa = 3.5RR101 pKa = 11.84HH102 pKa = 4.55GTAVSGVIAAALNNGVGGAGVAPGASLVGYY132 pKa = 10.17RR133 pKa = 11.84IGFGANGTLEE143 pKa = 4.01QLVAAFQLLTAVDD156 pKa = 3.8VANNSWGFDD165 pKa = 3.23GFFGDD170 pKa = 5.32NFLDD174 pKa = 3.92PDD176 pKa = 4.39FAPIGDD182 pKa = 4.29ALATALAAGRR192 pKa = 11.84GGLGTIVVMAAGNARR207 pKa = 11.84TSGQDD212 pKa = 3.2VNYY215 pKa = 10.56HH216 pKa = 6.04GFQNHH221 pKa = 6.65RR222 pKa = 11.84GTIAVAATDD231 pKa = 3.12SGGNVTYY238 pKa = 10.87YY239 pKa = 9.08STPGAALLVAAPGHH253 pKa = 6.59GITTTDD259 pKa = 3.13RR260 pKa = 11.84VDD262 pKa = 3.29GAGYY266 pKa = 11.0ASGDD270 pKa = 3.7YY271 pKa = 9.25ATLNGTSFAAPMVSGIAALLLDD293 pKa = 4.93ANPGLGWRR301 pKa = 11.84DD302 pKa = 3.42VQEE305 pKa = 4.15ILAATAVRR313 pKa = 11.84TGSPASWSFNAADD326 pKa = 3.44NWNGGGMHH334 pKa = 6.65VSHH337 pKa = 8.04DD338 pKa = 3.9YY339 pKa = 11.64GFGLVDD345 pKa = 4.02AYY347 pKa = 10.94AAVRR351 pKa = 11.84VAEE354 pKa = 4.31SWRR357 pKa = 11.84SVSTSLNEE365 pKa = 3.74WVAEE369 pKa = 4.02GLQYY373 pKa = 10.18PASPIAIPDD382 pKa = 3.85GGSASSTITLAAGLRR397 pKa = 11.84IDD399 pKa = 4.02RR400 pKa = 11.84VEE402 pKa = 3.78VDD404 pKa = 3.76LALAHH409 pKa = 6.93PYY411 pKa = 9.86LVQLRR416 pKa = 11.84VTLTAPDD423 pKa = 3.66GTEE426 pKa = 3.77SVLVQNPSTSQGNIYY441 pKa = 8.94FTFSTTRR448 pKa = 11.84DD449 pKa = 2.93WGEE452 pKa = 3.61FSGGNWTLTVTDD464 pKa = 4.05MQVGATGVVYY474 pKa = 10.27AWGIRR479 pKa = 11.84AYY481 pKa = 10.67GDD483 pKa = 3.39LAGDD487 pKa = 3.54DD488 pKa = 3.9TYY490 pKa = 11.74LYY492 pKa = 9.57TGEE495 pKa = 4.12FAALSAADD503 pKa = 3.32ASRR506 pKa = 11.84RR507 pKa = 11.84VLSDD511 pKa = 3.02AGGMDD516 pKa = 5.56AINAAAIAGDD526 pKa = 3.79TLLDD530 pKa = 3.6LRR532 pKa = 11.84PAHH535 pKa = 6.39VSLIAGQEE543 pKa = 4.0VTISAGTIIEE553 pKa = 4.27NADD556 pKa = 3.56SGDD559 pKa = 3.96GNDD562 pKa = 3.72TLIGNDD568 pKa = 3.33AANSLRR574 pKa = 11.84GWRR577 pKa = 11.84GNDD580 pKa = 3.35FLDD583 pKa = 4.29GGAGVDD589 pKa = 4.03TLDD592 pKa = 4.07GGAGVDD598 pKa = 3.76TLDD601 pKa = 4.23GGVGDD606 pKa = 4.55DD607 pKa = 3.9VYY609 pKa = 11.62VVDD612 pKa = 4.21VAADD616 pKa = 3.82VIVEE620 pKa = 4.16RR621 pKa = 11.84PGGGTDD627 pKa = 3.39TVRR630 pKa = 11.84TTLASYY636 pKa = 11.29LLGLEE641 pKa = 4.3LEE643 pKa = 4.34NLVFVGSGNFKK654 pKa = 9.67GTGNAAANVIDD665 pKa = 4.68GGAGNDD671 pKa = 3.8SLNGGLGADD680 pKa = 4.6LLRR683 pKa = 11.84GGLGDD688 pKa = 3.64DD689 pKa = 4.39TYY691 pKa = 11.24TVDD694 pKa = 3.14HH695 pKa = 7.0AGDD698 pKa = 3.98SVVEE702 pKa = 4.36LPGEE706 pKa = 4.39GNDD709 pKa = 3.64YY710 pKa = 10.98VYY712 pKa = 11.37SSVSWTLGANLEE724 pKa = 4.06RR725 pKa = 11.84LYY727 pKa = 10.39LTGSAAIDD735 pKa = 3.75GAGNDD740 pKa = 4.03LGNRR744 pKa = 11.84LYY746 pKa = 10.83GQSNSAINTLAGGPGNDD763 pKa = 3.31TYY765 pKa = 11.87YY766 pKa = 10.99VGSKK770 pKa = 10.7DD771 pKa = 4.43VIVEE775 pKa = 4.02LAGEE779 pKa = 4.52GTDD782 pKa = 3.25TAYY785 pKa = 10.49GYY787 pKa = 11.65GDD789 pKa = 3.56YY790 pKa = 10.46TLAAGVSVEE799 pKa = 3.78YY800 pKa = 9.77FYY802 pKa = 11.2INVTTGHH809 pKa = 6.03TLAGNEE815 pKa = 4.18LANNLRR821 pKa = 11.84GNSGNDD827 pKa = 3.21TLIGFEE833 pKa = 5.11GNDD836 pKa = 3.7SLNGGLGVDD845 pKa = 4.7LLRR848 pKa = 11.84GGPGDD853 pKa = 3.55DD854 pKa = 4.06TYY856 pKa = 11.5TVDD859 pKa = 3.14HH860 pKa = 7.0AGDD863 pKa = 3.9SVVEE867 pKa = 3.85LLGEE871 pKa = 4.51GKK873 pKa = 8.7DD874 pKa = 3.65TVYY877 pKa = 11.28SSVSWTLGDD886 pKa = 3.55HH887 pKa = 7.32LEE889 pKa = 4.03RR890 pKa = 11.84LYY892 pKa = 10.68LTGNAAIAGAGNEE905 pKa = 4.2LANTLVGYY913 pKa = 8.77TNAAGNALAGGAGDD927 pKa = 3.6DD928 pKa = 4.47AYY930 pKa = 11.5YY931 pKa = 11.16VDD933 pKa = 4.62ANDD936 pKa = 3.58VVVEE940 pKa = 4.04LVGEE944 pKa = 4.48GNDD947 pKa = 3.04IVYY950 pKa = 10.67GSVSWTLGANLEE962 pKa = 4.06RR963 pKa = 11.84LYY965 pKa = 10.39LTGSAAIDD973 pKa = 3.63GTGNDD978 pKa = 4.24LDD980 pKa = 3.72NRR982 pKa = 11.84LYY984 pKa = 10.93GQANGAINTLTGGTGNDD1001 pKa = 2.95IYY1003 pKa = 11.52YY1004 pKa = 10.53VGSNDD1009 pKa = 3.94VIVEE1013 pKa = 4.01LAVEE1017 pKa = 4.53GTDD1020 pKa = 3.16TAYY1023 pKa = 10.48GYY1025 pKa = 11.59GDD1027 pKa = 3.52YY1028 pKa = 10.66TLATGVSVEE1037 pKa = 3.9NLYY1040 pKa = 11.42LNVTTGQTLTGNEE1053 pKa = 4.32LANKK1057 pKa = 10.19LSGNAGSDD1065 pKa = 3.42TLRR1068 pKa = 11.84GLDD1071 pKa = 4.14GNDD1074 pKa = 3.46SLSGGLGADD1083 pKa = 3.76VLDD1086 pKa = 4.66GGQGNDD1092 pKa = 3.41TLAGGLGNDD1101 pKa = 3.83TVTGGNGNDD1110 pKa = 2.94IFRR1113 pKa = 11.84FATALDD1119 pKa = 4.02ANSNLDD1125 pKa = 3.43SVIDD1129 pKa = 3.82FNVVDD1134 pKa = 6.44DD1135 pKa = 5.08SFQLEE1140 pKa = 4.13NGIFTSLTQTGTLAVGLFVIGTAALDD1166 pKa = 4.08ANDD1169 pKa = 4.0KK1170 pKa = 10.85LIYY1173 pKa = 10.68DD1174 pKa = 4.02NTTGALFYY1182 pKa = 10.91DD1183 pKa = 4.5LDD1185 pKa = 4.36GSGSGGAIQFAVLSTNLALTNLDD1208 pKa = 3.86FVVTT1212 pKa = 4.39
MM1 pKa = 7.78ASFSPNMPTTANGRR15 pKa = 11.84TVTSQGVYY23 pKa = 10.28SDD25 pKa = 4.81PLLPSYY31 pKa = 9.91WYY33 pKa = 10.92LGDD36 pKa = 3.56ASGAVKK42 pKa = 10.28GVNAMRR48 pKa = 11.84AWDD51 pKa = 4.24DD52 pKa = 3.49YY53 pKa = 10.08RR54 pKa = 11.84GSGIVVAVIDD64 pKa = 4.51DD65 pKa = 4.03GVEE68 pKa = 4.03YY69 pKa = 9.66THH71 pKa = 7.43LDD73 pKa = 2.93LAANYY78 pKa = 10.01RR79 pKa = 11.84SDD81 pKa = 3.38LAYY84 pKa = 8.8DD85 pKa = 3.53TRR87 pKa = 11.84DD88 pKa = 3.21RR89 pKa = 11.84DD90 pKa = 3.33ADD92 pKa = 3.75AFPGEE97 pKa = 4.17SSDD100 pKa = 3.5RR101 pKa = 11.84HH102 pKa = 4.55GTAVSGVIAAALNNGVGGAGVAPGASLVGYY132 pKa = 10.17RR133 pKa = 11.84IGFGANGTLEE143 pKa = 4.01QLVAAFQLLTAVDD156 pKa = 3.8VANNSWGFDD165 pKa = 3.23GFFGDD170 pKa = 5.32NFLDD174 pKa = 3.92PDD176 pKa = 4.39FAPIGDD182 pKa = 4.29ALATALAAGRR192 pKa = 11.84GGLGTIVVMAAGNARR207 pKa = 11.84TSGQDD212 pKa = 3.2VNYY215 pKa = 10.56HH216 pKa = 6.04GFQNHH221 pKa = 6.65RR222 pKa = 11.84GTIAVAATDD231 pKa = 3.12SGGNVTYY238 pKa = 10.87YY239 pKa = 9.08STPGAALLVAAPGHH253 pKa = 6.59GITTTDD259 pKa = 3.13RR260 pKa = 11.84VDD262 pKa = 3.29GAGYY266 pKa = 11.0ASGDD270 pKa = 3.7YY271 pKa = 9.25ATLNGTSFAAPMVSGIAALLLDD293 pKa = 4.93ANPGLGWRR301 pKa = 11.84DD302 pKa = 3.42VQEE305 pKa = 4.15ILAATAVRR313 pKa = 11.84TGSPASWSFNAADD326 pKa = 3.44NWNGGGMHH334 pKa = 6.65VSHH337 pKa = 8.04DD338 pKa = 3.9YY339 pKa = 11.64GFGLVDD345 pKa = 4.02AYY347 pKa = 10.94AAVRR351 pKa = 11.84VAEE354 pKa = 4.31SWRR357 pKa = 11.84SVSTSLNEE365 pKa = 3.74WVAEE369 pKa = 4.02GLQYY373 pKa = 10.18PASPIAIPDD382 pKa = 3.85GGSASSTITLAAGLRR397 pKa = 11.84IDD399 pKa = 4.02RR400 pKa = 11.84VEE402 pKa = 3.78VDD404 pKa = 3.76LALAHH409 pKa = 6.93PYY411 pKa = 9.86LVQLRR416 pKa = 11.84VTLTAPDD423 pKa = 3.66GTEE426 pKa = 3.77SVLVQNPSTSQGNIYY441 pKa = 8.94FTFSTTRR448 pKa = 11.84DD449 pKa = 2.93WGEE452 pKa = 3.61FSGGNWTLTVTDD464 pKa = 4.05MQVGATGVVYY474 pKa = 10.27AWGIRR479 pKa = 11.84AYY481 pKa = 10.67GDD483 pKa = 3.39LAGDD487 pKa = 3.54DD488 pKa = 3.9TYY490 pKa = 11.74LYY492 pKa = 9.57TGEE495 pKa = 4.12FAALSAADD503 pKa = 3.32ASRR506 pKa = 11.84RR507 pKa = 11.84VLSDD511 pKa = 3.02AGGMDD516 pKa = 5.56AINAAAIAGDD526 pKa = 3.79TLLDD530 pKa = 3.6LRR532 pKa = 11.84PAHH535 pKa = 6.39VSLIAGQEE543 pKa = 4.0VTISAGTIIEE553 pKa = 4.27NADD556 pKa = 3.56SGDD559 pKa = 3.96GNDD562 pKa = 3.72TLIGNDD568 pKa = 3.33AANSLRR574 pKa = 11.84GWRR577 pKa = 11.84GNDD580 pKa = 3.35FLDD583 pKa = 4.29GGAGVDD589 pKa = 4.03TLDD592 pKa = 4.07GGAGVDD598 pKa = 3.76TLDD601 pKa = 4.23GGVGDD606 pKa = 4.55DD607 pKa = 3.9VYY609 pKa = 11.62VVDD612 pKa = 4.21VAADD616 pKa = 3.82VIVEE620 pKa = 4.16RR621 pKa = 11.84PGGGTDD627 pKa = 3.39TVRR630 pKa = 11.84TTLASYY636 pKa = 11.29LLGLEE641 pKa = 4.3LEE643 pKa = 4.34NLVFVGSGNFKK654 pKa = 9.67GTGNAAANVIDD665 pKa = 4.68GGAGNDD671 pKa = 3.8SLNGGLGADD680 pKa = 4.6LLRR683 pKa = 11.84GGLGDD688 pKa = 3.64DD689 pKa = 4.39TYY691 pKa = 11.24TVDD694 pKa = 3.14HH695 pKa = 7.0AGDD698 pKa = 3.98SVVEE702 pKa = 4.36LPGEE706 pKa = 4.39GNDD709 pKa = 3.64YY710 pKa = 10.98VYY712 pKa = 11.37SSVSWTLGANLEE724 pKa = 4.06RR725 pKa = 11.84LYY727 pKa = 10.39LTGSAAIDD735 pKa = 3.75GAGNDD740 pKa = 4.03LGNRR744 pKa = 11.84LYY746 pKa = 10.83GQSNSAINTLAGGPGNDD763 pKa = 3.31TYY765 pKa = 11.87YY766 pKa = 10.99VGSKK770 pKa = 10.7DD771 pKa = 4.43VIVEE775 pKa = 4.02LAGEE779 pKa = 4.52GTDD782 pKa = 3.25TAYY785 pKa = 10.49GYY787 pKa = 11.65GDD789 pKa = 3.56YY790 pKa = 10.46TLAAGVSVEE799 pKa = 3.78YY800 pKa = 9.77FYY802 pKa = 11.2INVTTGHH809 pKa = 6.03TLAGNEE815 pKa = 4.18LANNLRR821 pKa = 11.84GNSGNDD827 pKa = 3.21TLIGFEE833 pKa = 5.11GNDD836 pKa = 3.7SLNGGLGVDD845 pKa = 4.7LLRR848 pKa = 11.84GGPGDD853 pKa = 3.55DD854 pKa = 4.06TYY856 pKa = 11.5TVDD859 pKa = 3.14HH860 pKa = 7.0AGDD863 pKa = 3.9SVVEE867 pKa = 3.85LLGEE871 pKa = 4.51GKK873 pKa = 8.7DD874 pKa = 3.65TVYY877 pKa = 11.28SSVSWTLGDD886 pKa = 3.55HH887 pKa = 7.32LEE889 pKa = 4.03RR890 pKa = 11.84LYY892 pKa = 10.68LTGNAAIAGAGNEE905 pKa = 4.2LANTLVGYY913 pKa = 8.77TNAAGNALAGGAGDD927 pKa = 3.6DD928 pKa = 4.47AYY930 pKa = 11.5YY931 pKa = 11.16VDD933 pKa = 4.62ANDD936 pKa = 3.58VVVEE940 pKa = 4.04LVGEE944 pKa = 4.48GNDD947 pKa = 3.04IVYY950 pKa = 10.67GSVSWTLGANLEE962 pKa = 4.06RR963 pKa = 11.84LYY965 pKa = 10.39LTGSAAIDD973 pKa = 3.63GTGNDD978 pKa = 4.24LDD980 pKa = 3.72NRR982 pKa = 11.84LYY984 pKa = 10.93GQANGAINTLTGGTGNDD1001 pKa = 2.95IYY1003 pKa = 11.52YY1004 pKa = 10.53VGSNDD1009 pKa = 3.94VIVEE1013 pKa = 4.01LAVEE1017 pKa = 4.53GTDD1020 pKa = 3.16TAYY1023 pKa = 10.48GYY1025 pKa = 11.59GDD1027 pKa = 3.52YY1028 pKa = 10.66TLATGVSVEE1037 pKa = 3.9NLYY1040 pKa = 11.42LNVTTGQTLTGNEE1053 pKa = 4.32LANKK1057 pKa = 10.19LSGNAGSDD1065 pKa = 3.42TLRR1068 pKa = 11.84GLDD1071 pKa = 4.14GNDD1074 pKa = 3.46SLSGGLGADD1083 pKa = 3.76VLDD1086 pKa = 4.66GGQGNDD1092 pKa = 3.41TLAGGLGNDD1101 pKa = 3.83TVTGGNGNDD1110 pKa = 2.94IFRR1113 pKa = 11.84FATALDD1119 pKa = 4.02ANSNLDD1125 pKa = 3.43SVIDD1129 pKa = 3.82FNVVDD1134 pKa = 6.44DD1135 pKa = 5.08SFQLEE1140 pKa = 4.13NGIFTSLTQTGTLAVGLFVIGTAALDD1166 pKa = 4.08ANDD1169 pKa = 4.0KK1170 pKa = 10.85LIYY1173 pKa = 10.68DD1174 pKa = 4.02NTTGALFYY1182 pKa = 10.91DD1183 pKa = 4.5LDD1185 pKa = 4.36GSGSGGAIQFAVLSTNLALTNLDD1208 pKa = 3.86FVVTT1212 pKa = 4.39
Molecular weight: 123.63 kDa
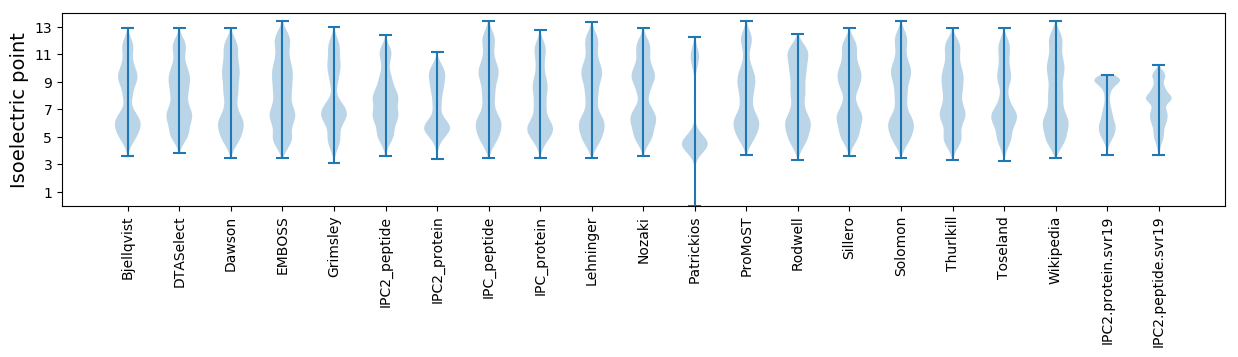
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A011P3T9|A0A011P3T9_9PROT OsmC-like protein OS=Candidatus Accumulibacter sp. SK-11 OX=1454000 GN=AW07_03572 PE=4 SV=1
MM1 pKa = 7.37SAITRR6 pKa = 11.84MSVFGSLAAVSIIRR20 pKa = 11.84SAHH23 pKa = 4.39IVARR27 pKa = 11.84FRR29 pKa = 11.84SALPAAIRR37 pKa = 11.84RR38 pKa = 11.84PTRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FSMTASRR50 pKa = 11.84SMIGIAHH57 pKa = 6.9SSPSFSAVTVV67 pKa = 3.6
MM1 pKa = 7.37SAITRR6 pKa = 11.84MSVFGSLAAVSIIRR20 pKa = 11.84SAHH23 pKa = 4.39IVARR27 pKa = 11.84FRR29 pKa = 11.84SALPAAIRR37 pKa = 11.84RR38 pKa = 11.84PTRR41 pKa = 11.84RR42 pKa = 11.84RR43 pKa = 11.84FSMTASRR50 pKa = 11.84SMIGIAHH57 pKa = 6.9SSPSFSAVTVV67 pKa = 3.6
Molecular weight: 7.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1187205 |
29 |
4072 |
249.0 |
27.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.706 ± 0.059 | 1.247 ± 0.015 |
5.359 ± 0.033 | 5.459 ± 0.04 |
3.319 ± 0.022 | 8.241 ± 0.044 |
2.329 ± 0.018 | 4.325 ± 0.026 |
2.526 ± 0.035 | 10.995 ± 0.054 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.15 ± 0.017 | 2.297 ± 0.024 |
5.296 ± 0.033 | 3.977 ± 0.027 |
8.624 ± 0.047 | 5.718 ± 0.039 |
4.582 ± 0.028 | 7.392 ± 0.046 |
1.341 ± 0.016 | 1.967 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
