
Duwamo virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 6.78
Get precalculated fractions of proteins
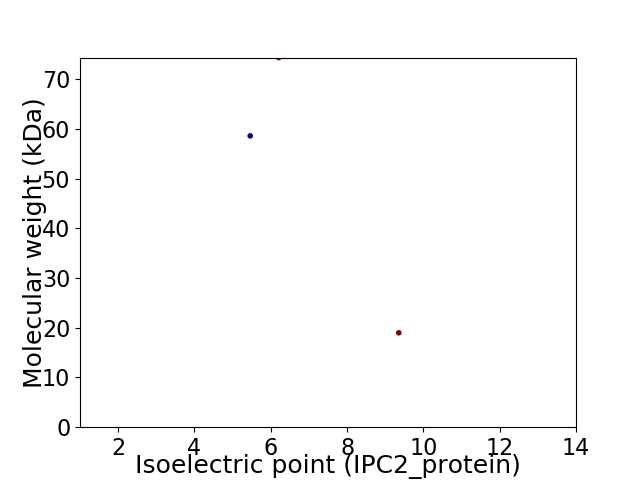
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1B2RVQ1|A0A1B2RVQ1_9VIRU Capsid protein OS=Duwamo virus OX=1888317 PE=4 SV=1
MM1 pKa = 7.71SSTSHH6 pKa = 6.61IPQPAAGATDD16 pKa = 3.7EE17 pKa = 5.16VFDD20 pKa = 4.19STSATAVQPTVSRR33 pKa = 11.84LGIPAGPPQEE43 pKa = 4.64IPIHH47 pKa = 5.42TGQVNTYY54 pKa = 9.23DD55 pKa = 4.57ASIHH59 pKa = 4.92QQWVPTDD66 pKa = 3.95SITWTVSQPSGTLLWFKK83 pKa = 10.09PIHH86 pKa = 5.59PTFSNALLAYY96 pKa = 10.37LSRR99 pKa = 11.84IYY101 pKa = 10.37NAWGGALDD109 pKa = 4.15YY110 pKa = 10.53KK111 pKa = 10.65FKK113 pKa = 10.69IAGTGFHH120 pKa = 7.02AGAIAIVRR128 pKa = 11.84IPPNRR133 pKa = 11.84NPSEE137 pKa = 4.03FTTPQSWGAFEE148 pKa = 5.05YY149 pKa = 10.37MVIDD153 pKa = 4.45PKK155 pKa = 10.62TLEE158 pKa = 4.49TISVGISDD166 pKa = 4.17QRR168 pKa = 11.84PIAYY172 pKa = 9.48HH173 pKa = 4.93YY174 pKa = 10.51FPYY177 pKa = 10.62NGEE180 pKa = 4.12NPLSFGGWIAMYY192 pKa = 10.6VLIPLNTSSSGSQTISIQSFCKK214 pKa = 9.42PAEE217 pKa = 3.96FFQYY221 pKa = 10.37SQLIMPSEE229 pKa = 4.69NISADD234 pKa = 3.51PFPSEE239 pKa = 3.55FAYY242 pKa = 10.51AFDD245 pKa = 3.65FTTAKK250 pKa = 10.38YY251 pKa = 10.26CATAPLYY258 pKa = 10.42CDD260 pKa = 3.39QVTLEE265 pKa = 4.67ANSVTQSFYY274 pKa = 11.39VANCYY279 pKa = 8.78STGGDD284 pKa = 4.0RR285 pKa = 11.84LTKK288 pKa = 9.86WMSKK292 pKa = 9.33GSKK295 pKa = 10.27FPDD298 pKa = 3.04VRR300 pKa = 11.84DD301 pKa = 3.44THH303 pKa = 8.02RR304 pKa = 11.84EE305 pKa = 3.52GKK307 pKa = 7.36TQMRR311 pKa = 11.84ALTDD315 pKa = 3.96LSDD318 pKa = 3.41EE319 pKa = 4.44TIKK322 pKa = 11.14VKK324 pKa = 10.71LYY326 pKa = 10.06FKK328 pKa = 10.53QDD330 pKa = 3.6YY331 pKa = 10.35YY332 pKa = 10.64PIALPASKK340 pKa = 10.36EE341 pKa = 4.18NYY343 pKa = 9.58VSLLTYY349 pKa = 10.53SSEE352 pKa = 4.21NVKK355 pKa = 10.79NEE357 pKa = 3.53AWHH360 pKa = 5.7EE361 pKa = 4.26RR362 pKa = 11.84TWTALYY368 pKa = 10.88SEE370 pKa = 5.92DD371 pKa = 3.75NVAWQCATAATDD383 pKa = 3.26IVTDD387 pKa = 3.79TVVSIQNAKK396 pKa = 10.08FVLDD400 pKa = 4.04PSYY403 pKa = 11.37TDD405 pKa = 3.3NTFSTAVTGEE415 pKa = 4.47SILHH419 pKa = 5.82FEE421 pKa = 4.14NTISKK426 pKa = 9.79VKK428 pKa = 10.42SIQTEE433 pKa = 3.75EE434 pKa = 3.8MAALFSTGRR443 pKa = 11.84VKK445 pKa = 10.45QAFYY449 pKa = 11.52GNMCVLFTMISVDD462 pKa = 3.31EE463 pKa = 4.76DD464 pKa = 4.01LPIGYY469 pKa = 9.97AKK471 pKa = 10.51LYY473 pKa = 10.5QEE475 pKa = 4.99GFLSIRR481 pKa = 11.84VGNKK485 pKa = 7.52QVVLKK490 pKa = 10.07PSNLRR495 pKa = 11.84LKK497 pKa = 10.69FSGFIARR504 pKa = 11.84TDD506 pKa = 4.86PIPSSAEE513 pKa = 3.83YY514 pKa = 9.69NKK516 pKa = 10.31NMYY519 pKa = 8.42MVRR522 pKa = 11.84ALSRR526 pKa = 11.84NN527 pKa = 3.51
MM1 pKa = 7.71SSTSHH6 pKa = 6.61IPQPAAGATDD16 pKa = 3.7EE17 pKa = 5.16VFDD20 pKa = 4.19STSATAVQPTVSRR33 pKa = 11.84LGIPAGPPQEE43 pKa = 4.64IPIHH47 pKa = 5.42TGQVNTYY54 pKa = 9.23DD55 pKa = 4.57ASIHH59 pKa = 4.92QQWVPTDD66 pKa = 3.95SITWTVSQPSGTLLWFKK83 pKa = 10.09PIHH86 pKa = 5.59PTFSNALLAYY96 pKa = 10.37LSRR99 pKa = 11.84IYY101 pKa = 10.37NAWGGALDD109 pKa = 4.15YY110 pKa = 10.53KK111 pKa = 10.65FKK113 pKa = 10.69IAGTGFHH120 pKa = 7.02AGAIAIVRR128 pKa = 11.84IPPNRR133 pKa = 11.84NPSEE137 pKa = 4.03FTTPQSWGAFEE148 pKa = 5.05YY149 pKa = 10.37MVIDD153 pKa = 4.45PKK155 pKa = 10.62TLEE158 pKa = 4.49TISVGISDD166 pKa = 4.17QRR168 pKa = 11.84PIAYY172 pKa = 9.48HH173 pKa = 4.93YY174 pKa = 10.51FPYY177 pKa = 10.62NGEE180 pKa = 4.12NPLSFGGWIAMYY192 pKa = 10.6VLIPLNTSSSGSQTISIQSFCKK214 pKa = 9.42PAEE217 pKa = 3.96FFQYY221 pKa = 10.37SQLIMPSEE229 pKa = 4.69NISADD234 pKa = 3.51PFPSEE239 pKa = 3.55FAYY242 pKa = 10.51AFDD245 pKa = 3.65FTTAKK250 pKa = 10.38YY251 pKa = 10.26CATAPLYY258 pKa = 10.42CDD260 pKa = 3.39QVTLEE265 pKa = 4.67ANSVTQSFYY274 pKa = 11.39VANCYY279 pKa = 8.78STGGDD284 pKa = 4.0RR285 pKa = 11.84LTKK288 pKa = 9.86WMSKK292 pKa = 9.33GSKK295 pKa = 10.27FPDD298 pKa = 3.04VRR300 pKa = 11.84DD301 pKa = 3.44THH303 pKa = 8.02RR304 pKa = 11.84EE305 pKa = 3.52GKK307 pKa = 7.36TQMRR311 pKa = 11.84ALTDD315 pKa = 3.96LSDD318 pKa = 3.41EE319 pKa = 4.44TIKK322 pKa = 11.14VKK324 pKa = 10.71LYY326 pKa = 10.06FKK328 pKa = 10.53QDD330 pKa = 3.6YY331 pKa = 10.35YY332 pKa = 10.64PIALPASKK340 pKa = 10.36EE341 pKa = 4.18NYY343 pKa = 9.58VSLLTYY349 pKa = 10.53SSEE352 pKa = 4.21NVKK355 pKa = 10.79NEE357 pKa = 3.53AWHH360 pKa = 5.7EE361 pKa = 4.26RR362 pKa = 11.84TWTALYY368 pKa = 10.88SEE370 pKa = 5.92DD371 pKa = 3.75NVAWQCATAATDD383 pKa = 3.26IVTDD387 pKa = 3.79TVVSIQNAKK396 pKa = 10.08FVLDD400 pKa = 4.04PSYY403 pKa = 11.37TDD405 pKa = 3.3NTFSTAVTGEE415 pKa = 4.47SILHH419 pKa = 5.82FEE421 pKa = 4.14NTISKK426 pKa = 9.79VKK428 pKa = 10.42SIQTEE433 pKa = 3.75EE434 pKa = 3.8MAALFSTGRR443 pKa = 11.84VKK445 pKa = 10.45QAFYY449 pKa = 11.52GNMCVLFTMISVDD462 pKa = 3.31EE463 pKa = 4.76DD464 pKa = 4.01LPIGYY469 pKa = 9.97AKK471 pKa = 10.51LYY473 pKa = 10.5QEE475 pKa = 4.99GFLSIRR481 pKa = 11.84VGNKK485 pKa = 7.52QVVLKK490 pKa = 10.07PSNLRR495 pKa = 11.84LKK497 pKa = 10.69FSGFIARR504 pKa = 11.84TDD506 pKa = 4.86PIPSSAEE513 pKa = 3.83YY514 pKa = 9.69NKK516 pKa = 10.31NMYY519 pKa = 8.42MVRR522 pKa = 11.84ALSRR526 pKa = 11.84NN527 pKa = 3.51
Molecular weight: 58.6 kDa
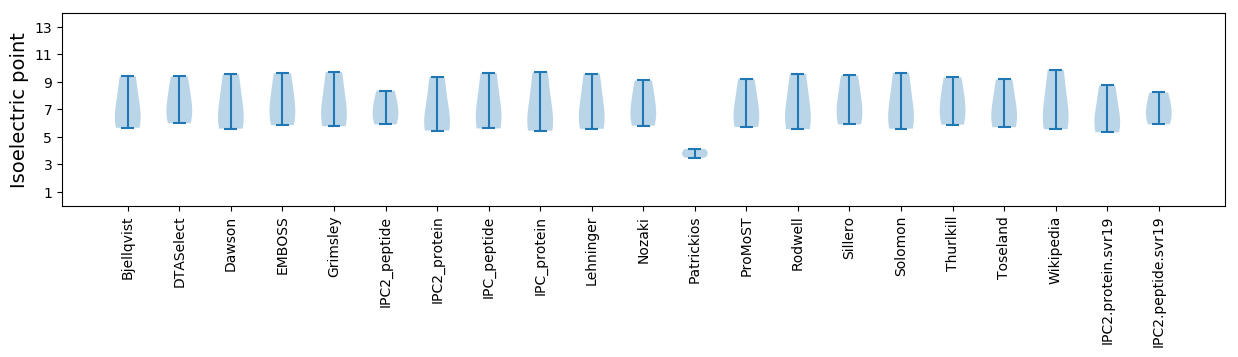
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1B2RVP9|A0A1B2RVP9_9VIRU RNA-dependent RNA polymerase OS=Duwamo virus OX=1888317 PE=4 SV=1
MM1 pKa = 7.28AAAAIGATAAAGASNIVSSGLTAGLQIYY29 pKa = 10.13GNSLNNSQQFAYY41 pKa = 9.57NDD43 pKa = 3.9SVISRR48 pKa = 11.84AEE50 pKa = 3.69HH51 pKa = 6.58AFTDD55 pKa = 3.65NGLPKK60 pKa = 10.71YY61 pKa = 9.25MAYY64 pKa = 10.04QSNSGTTPTKK74 pKa = 9.87MYY76 pKa = 10.14QISGGNFYY84 pKa = 10.99SAGPVNSNLPMYY96 pKa = 7.65TTTAQQAFHH105 pKa = 6.88AAAPRR110 pKa = 11.84PNARR114 pKa = 11.84VGPGEE119 pKa = 3.76PAIPNWGDD127 pKa = 2.9IHH129 pKa = 8.12NGMPMVPVGQNNRR142 pKa = 11.84QGLGFGRR149 pKa = 11.84YY150 pKa = 8.94NIANNFVQGGPMLNANVRR168 pKa = 11.84NADD171 pKa = 3.19QHH173 pKa = 5.18QAFARR178 pKa = 11.84ALQQ181 pKa = 3.59
MM1 pKa = 7.28AAAAIGATAAAGASNIVSSGLTAGLQIYY29 pKa = 10.13GNSLNNSQQFAYY41 pKa = 9.57NDD43 pKa = 3.9SVISRR48 pKa = 11.84AEE50 pKa = 3.69HH51 pKa = 6.58AFTDD55 pKa = 3.65NGLPKK60 pKa = 10.71YY61 pKa = 9.25MAYY64 pKa = 10.04QSNSGTTPTKK74 pKa = 9.87MYY76 pKa = 10.14QISGGNFYY84 pKa = 10.99SAGPVNSNLPMYY96 pKa = 7.65TTTAQQAFHH105 pKa = 6.88AAAPRR110 pKa = 11.84PNARR114 pKa = 11.84VGPGEE119 pKa = 3.76PAIPNWGDD127 pKa = 2.9IHH129 pKa = 8.12NGMPMVPVGQNNRR142 pKa = 11.84QGLGFGRR149 pKa = 11.84YY150 pKa = 8.94NIANNFVQGGPMLNANVRR168 pKa = 11.84NADD171 pKa = 3.19QHH173 pKa = 5.18QAFARR178 pKa = 11.84ALQQ181 pKa = 3.59
Molecular weight: 18.99 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1355 |
181 |
647 |
451.7 |
50.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.266 ± 1.57 | 1.033 ± 0.191 |
5.166 ± 0.822 | 4.723 ± 0.799 |
4.797 ± 0.228 | 6.052 ± 0.878 |
3.1 ± 0.821 | 5.609 ± 0.362 |
5.83 ± 1.434 | 7.675 ± 1.22 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.435 ± 0.251 | 5.387 ± 1.119 |
5.535 ± 0.527 | 4.133 ± 0.701 |
3.321 ± 0.127 | 7.159 ± 1.321 |
7.159 ± 0.675 | 5.978 ± 0.351 |
1.771 ± 0.225 | 4.871 ± 0.111 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
