
Xanthomonadaceae bacterium
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Xanthomonadales; Xanthomonadaceae; unclassified Xanthomonadaceae
Average proteome isoelectric point is 6.69
Get precalculated fractions of proteins
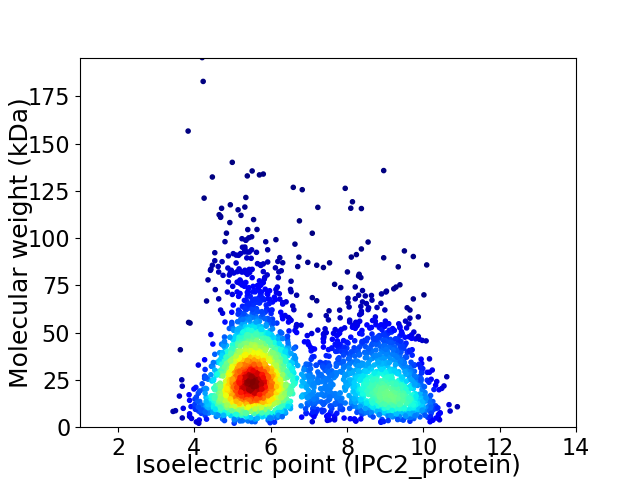
Virtual 2D-PAGE plot for 3169 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
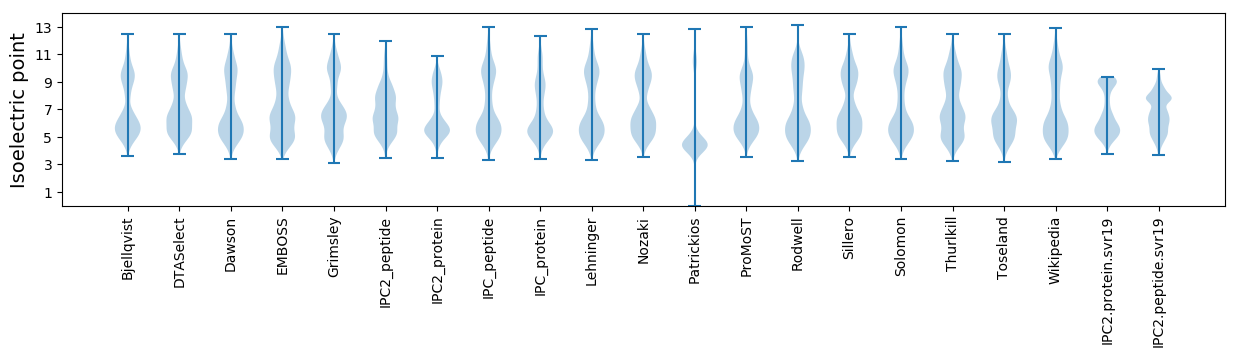
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4Q6C7C1|A0A4Q6C7C1_9GAMM TerC family protein (Fragment) OS=Xanthomonadaceae bacterium OX=1926873 GN=EOP91_07615 PE=4 SV=1
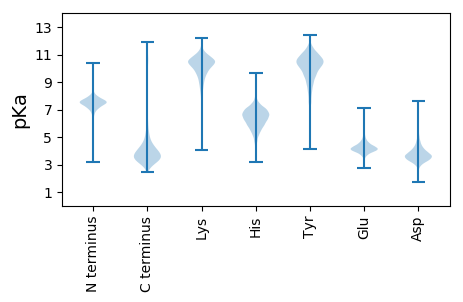
MM1 pKa = 7.68TDD3 pKa = 3.03STDD6 pKa = 3.15ARR8 pKa = 11.84YY9 pKa = 7.35QTWMCVVCGFIYY21 pKa = 10.67DD22 pKa = 3.87EE23 pKa = 4.55AKK25 pKa = 10.57GLPEE29 pKa = 4.53EE30 pKa = 4.68GLAPGTRR37 pKa = 11.84WKK39 pKa = 10.41DD40 pKa = 3.32IPDD43 pKa = 3.11SWTCPDD49 pKa = 4.16CGVGKK54 pKa = 10.25DD55 pKa = 3.81DD56 pKa = 4.93FEE58 pKa = 4.83MIEE61 pKa = 3.83VDD63 pKa = 3.41
MM1 pKa = 7.68TDD3 pKa = 3.03STDD6 pKa = 3.15ARR8 pKa = 11.84YY9 pKa = 7.35QTWMCVVCGFIYY21 pKa = 10.67DD22 pKa = 3.87EE23 pKa = 4.55AKK25 pKa = 10.57GLPEE29 pKa = 4.53EE30 pKa = 4.68GLAPGTRR37 pKa = 11.84WKK39 pKa = 10.41DD40 pKa = 3.32IPDD43 pKa = 3.11SWTCPDD49 pKa = 4.16CGVGKK54 pKa = 10.25DD55 pKa = 3.81DD56 pKa = 4.93FEE58 pKa = 4.83MIEE61 pKa = 3.83VDD63 pKa = 3.41
Molecular weight: 7.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4Q6C685|A0A4Q6C685_9GAMM Fused isobutyryl-CoA mutase OS=Xanthomonadaceae bacterium OX=1926873 GN=icmF PE=3 SV=1
MM1 pKa = 7.91LDD3 pKa = 3.7RR4 pKa = 11.84VRR6 pKa = 11.84DD7 pKa = 3.88CCRR10 pKa = 11.84VRR12 pKa = 11.84HH13 pKa = 5.04YY14 pKa = 10.96SLRR17 pKa = 11.84TEE19 pKa = 3.94RR20 pKa = 11.84AYY22 pKa = 11.33VDD24 pKa = 3.71WIRR27 pKa = 11.84RR28 pKa = 11.84FVVANDD34 pKa = 3.09KK35 pKa = 10.53RR36 pKa = 11.84HH37 pKa = 5.77PRR39 pKa = 11.84TMGAPEE45 pKa = 3.97VEE47 pKa = 4.55AFLSTLATTHH57 pKa = 7.0DD58 pKa = 4.12VAASTQNQALSALLFLYY75 pKa = 10.6RR76 pKa = 11.84EE77 pKa = 4.25VLGMQLPWMEE87 pKa = 4.0SVVRR91 pKa = 11.84AKK93 pKa = 10.44RR94 pKa = 11.84PQKK97 pKa = 10.32IPAVLSRR104 pKa = 11.84DD105 pKa = 3.35EE106 pKa = 4.22VGRR109 pKa = 11.84LLALMDD115 pKa = 3.85GVRR118 pKa = 11.84NGKK121 pKa = 9.27GGKK124 pKa = 8.65DD125 pKa = 2.79RR126 pKa = 11.84RR127 pKa = 11.84VPLPRR132 pKa = 11.84RR133 pKa = 11.84LRR135 pKa = 11.84EE136 pKa = 3.74RR137 pKa = 11.84LLEE140 pKa = 3.8QVEE143 pKa = 4.43RR144 pKa = 11.84VRR146 pKa = 11.84VQHH149 pKa = 6.38GRR151 pKa = 11.84DD152 pKa = 3.23LAGGAGEE159 pKa = 4.46VYY161 pKa = 10.39LPHH164 pKa = 7.06ALARR168 pKa = 11.84KK169 pKa = 8.69YY170 pKa = 10.56PNAGRR175 pKa = 11.84QFCWQYY181 pKa = 10.63VFPSPRR187 pKa = 11.84EE188 pKa = 3.84SRR190 pKa = 11.84DD191 pKa = 3.31PRR193 pKa = 11.84SGRR196 pKa = 11.84VRR198 pKa = 11.84RR199 pKa = 11.84HH200 pKa = 6.21HH201 pKa = 6.72IDD203 pKa = 3.06EE204 pKa = 4.85GVLQRR209 pKa = 11.84AVKK212 pKa = 9.77VARR215 pKa = 11.84DD216 pKa = 3.11RR217 pKa = 11.84AGIDD221 pKa = 3.43KK222 pKa = 9.86PASCHH227 pKa = 5.08TLRR230 pKa = 11.84HH231 pKa = 5.5SFATHH236 pKa = 6.51LLEE239 pKa = 4.74SGQDD243 pKa = 2.86IRR245 pKa = 11.84TVQEE249 pKa = 3.9LMGHH253 pKa = 6.42KK254 pKa = 10.07DD255 pKa = 3.11VSTTQIYY262 pKa = 7.47THH264 pKa = 5.38VLGRR268 pKa = 11.84GAHH271 pKa = 5.8GVLSPLDD278 pKa = 3.41QQ279 pKa = 4.9
MM1 pKa = 7.91LDD3 pKa = 3.7RR4 pKa = 11.84VRR6 pKa = 11.84DD7 pKa = 3.88CCRR10 pKa = 11.84VRR12 pKa = 11.84HH13 pKa = 5.04YY14 pKa = 10.96SLRR17 pKa = 11.84TEE19 pKa = 3.94RR20 pKa = 11.84AYY22 pKa = 11.33VDD24 pKa = 3.71WIRR27 pKa = 11.84RR28 pKa = 11.84FVVANDD34 pKa = 3.09KK35 pKa = 10.53RR36 pKa = 11.84HH37 pKa = 5.77PRR39 pKa = 11.84TMGAPEE45 pKa = 3.97VEE47 pKa = 4.55AFLSTLATTHH57 pKa = 7.0DD58 pKa = 4.12VAASTQNQALSALLFLYY75 pKa = 10.6RR76 pKa = 11.84EE77 pKa = 4.25VLGMQLPWMEE87 pKa = 4.0SVVRR91 pKa = 11.84AKK93 pKa = 10.44RR94 pKa = 11.84PQKK97 pKa = 10.32IPAVLSRR104 pKa = 11.84DD105 pKa = 3.35EE106 pKa = 4.22VGRR109 pKa = 11.84LLALMDD115 pKa = 3.85GVRR118 pKa = 11.84NGKK121 pKa = 9.27GGKK124 pKa = 8.65DD125 pKa = 2.79RR126 pKa = 11.84RR127 pKa = 11.84VPLPRR132 pKa = 11.84RR133 pKa = 11.84LRR135 pKa = 11.84EE136 pKa = 3.74RR137 pKa = 11.84LLEE140 pKa = 3.8QVEE143 pKa = 4.43RR144 pKa = 11.84VRR146 pKa = 11.84VQHH149 pKa = 6.38GRR151 pKa = 11.84DD152 pKa = 3.23LAGGAGEE159 pKa = 4.46VYY161 pKa = 10.39LPHH164 pKa = 7.06ALARR168 pKa = 11.84KK169 pKa = 8.69YY170 pKa = 10.56PNAGRR175 pKa = 11.84QFCWQYY181 pKa = 10.63VFPSPRR187 pKa = 11.84EE188 pKa = 3.84SRR190 pKa = 11.84DD191 pKa = 3.31PRR193 pKa = 11.84SGRR196 pKa = 11.84VRR198 pKa = 11.84RR199 pKa = 11.84HH200 pKa = 6.21HH201 pKa = 6.72IDD203 pKa = 3.06EE204 pKa = 4.85GVLQRR209 pKa = 11.84AVKK212 pKa = 9.77VARR215 pKa = 11.84DD216 pKa = 3.11RR217 pKa = 11.84AGIDD221 pKa = 3.43KK222 pKa = 9.86PASCHH227 pKa = 5.08TLRR230 pKa = 11.84HH231 pKa = 5.5SFATHH236 pKa = 6.51LLEE239 pKa = 4.74SGQDD243 pKa = 2.86IRR245 pKa = 11.84TVQEE249 pKa = 3.9LMGHH253 pKa = 6.42KK254 pKa = 10.07DD255 pKa = 3.11VSTTQIYY262 pKa = 7.47THH264 pKa = 5.38VLGRR268 pKa = 11.84GAHH271 pKa = 5.8GVLSPLDD278 pKa = 3.41QQ279 pKa = 4.9
Molecular weight: 31.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
897507 |
19 |
1915 |
283.2 |
30.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.033 ± 0.066 | 0.834 ± 0.014 |
5.666 ± 0.032 | 5.422 ± 0.043 |
3.411 ± 0.03 | 8.808 ± 0.044 |
2.199 ± 0.023 | 4.034 ± 0.035 |
2.76 ± 0.036 | 10.988 ± 0.056 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.181 ± 0.018 | 2.442 ± 0.03 |
5.467 ± 0.039 | 3.805 ± 0.029 |
7.673 ± 0.051 | 5.413 ± 0.032 |
4.588 ± 0.038 | 7.413 ± 0.034 |
1.544 ± 0.021 | 2.319 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
