
Magnaporthe oryzae RNA virus
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 8.57
Get precalculated fractions of proteins
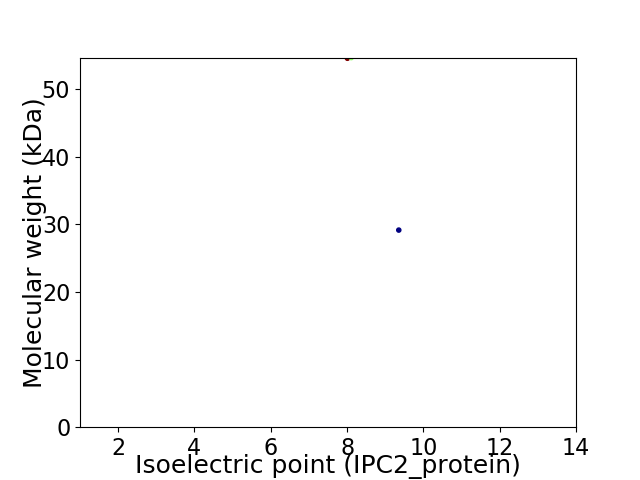
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
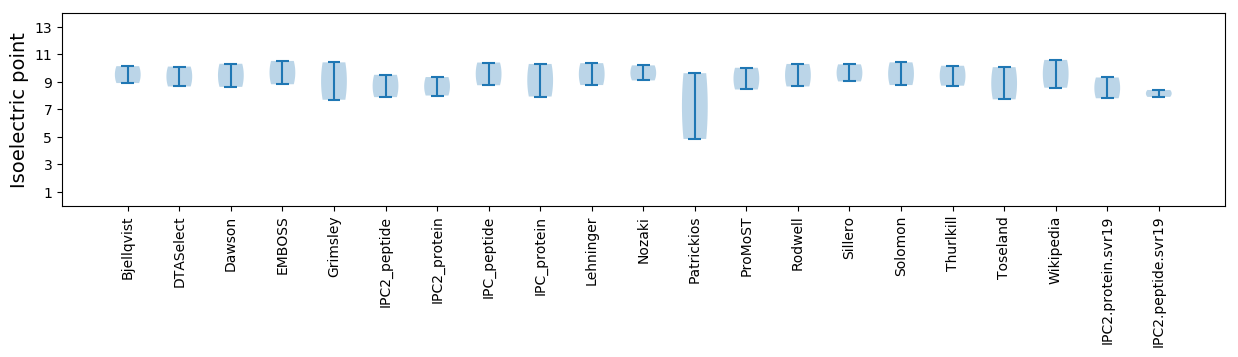
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7RYK7|A0A0A7RYK7_9VIRU RNA-dependent RNA polymerase OS=Magnaporthe oryzae RNA virus OX=1580607 GN=RdRp PE=4 SV=1
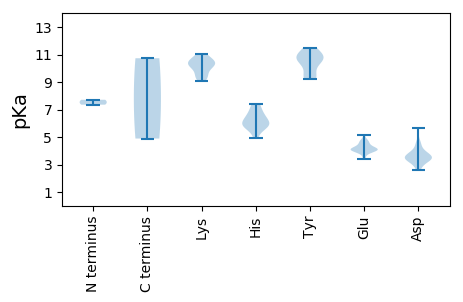
MM1 pKa = 7.35RR2 pKa = 11.84TACPTSFVGMWVPAVHH18 pKa = 7.25ADD20 pKa = 3.68CPHH23 pKa = 5.93NQVASLLKK31 pKa = 9.42RR32 pKa = 11.84TLGPVPDD39 pKa = 4.66PVHH42 pKa = 6.62EE43 pKa = 4.23PVGPTVKK50 pKa = 10.46AVFSRR55 pKa = 11.84LRR57 pKa = 11.84ALAARR62 pKa = 11.84SGVSRR67 pKa = 11.84WSLRR71 pKa = 11.84RR72 pKa = 11.84TAEE75 pKa = 4.06SYY77 pKa = 10.54KK78 pKa = 10.22GALGRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 9.19MEE87 pKa = 4.26AEE89 pKa = 3.45RR90 pKa = 11.84SLRR93 pKa = 11.84VDD95 pKa = 3.46GPVNSGDD102 pKa = 3.3TYY104 pKa = 11.36LRR106 pKa = 11.84PFLKK110 pKa = 10.59AEE112 pKa = 4.14KK113 pKa = 9.99VKK115 pKa = 11.01VMDD118 pKa = 4.0KK119 pKa = 10.3QSKK122 pKa = 9.27PRR124 pKa = 11.84MIFPRR129 pKa = 11.84SPRR132 pKa = 11.84YY133 pKa = 9.31NLSIASRR140 pKa = 11.84LKK142 pKa = 10.7PFEE145 pKa = 3.71HH146 pKa = 6.45WVWGRR151 pKa = 11.84LTTQVVLGTGVGRR164 pKa = 11.84VVAKK168 pKa = 10.32GLNPRR173 pKa = 11.84QRR175 pKa = 11.84ANLIVRR181 pKa = 11.84KK182 pKa = 9.49FRR184 pKa = 11.84ALDD187 pKa = 3.53DD188 pKa = 4.25CVCVEE193 pKa = 4.11VDD195 pKa = 3.6GKK197 pKa = 10.99AFEE200 pKa = 4.12AHH202 pKa = 5.87VGEE205 pKa = 4.52DD206 pKa = 3.39QLLEE210 pKa = 3.85EE211 pKa = 4.66HH212 pKa = 6.39GVYY215 pKa = 9.77EE216 pKa = 4.76AAFPGDD222 pKa = 3.43WEE224 pKa = 4.25LQRR227 pKa = 11.84LLRR230 pKa = 11.84VQLSLRR236 pKa = 11.84GKK238 pKa = 9.06LPCGAEE244 pKa = 4.05FTRR247 pKa = 11.84PGGRR251 pKa = 11.84ASGDD255 pKa = 3.49FNTGMGNSLIMLAVVGAVMKK275 pKa = 10.12EE276 pKa = 3.9LAPGAFDD283 pKa = 5.68LLVDD287 pKa = 4.48GDD289 pKa = 3.96NALVFMPRR297 pKa = 11.84GLLGLVRR304 pKa = 11.84ANFEE308 pKa = 4.03RR309 pKa = 11.84RR310 pKa = 11.84VLEE313 pKa = 4.06VSGHH317 pKa = 6.02EE318 pKa = 4.08IQLEE322 pKa = 4.58SPCSVIEE329 pKa = 4.24EE330 pKa = 4.08IRR332 pKa = 11.84FGQSAPINLGGSRR345 pKa = 11.84GWTMVRR351 pKa = 11.84DD352 pKa = 3.74PRR354 pKa = 11.84KK355 pKa = 10.33VISQALSSHH364 pKa = 5.72RR365 pKa = 11.84WLRR368 pKa = 11.84EE369 pKa = 3.41PTFASEE375 pKa = 4.74WIRR378 pKa = 11.84GVAACEE384 pKa = 3.99LSLSRR389 pKa = 11.84GVPVLQAWSASLQRR403 pKa = 11.84VYY405 pKa = 10.92GGPGGVRR412 pKa = 11.84EE413 pKa = 4.76HH414 pKa = 6.78PHH416 pKa = 5.64TDD418 pKa = 3.06YY419 pKa = 10.85FYY421 pKa = 10.59QGAWFADD428 pKa = 3.65VSQSVEE434 pKa = 3.86VTQQARR440 pKa = 11.84ISFWAAFGVAPDD452 pKa = 3.83EE453 pKa = 4.06QLALEE458 pKa = 4.81GGAFEE463 pKa = 5.13PSSEE467 pKa = 4.06DD468 pKa = 3.33YY469 pKa = 10.99EE470 pKa = 4.49RR471 pKa = 11.84VVLEE475 pKa = 4.01SFQWRR480 pKa = 11.84DD481 pKa = 3.42MPPGVAEE488 pKa = 4.41PLADD492 pKa = 3.67SLL494 pKa = 4.89
MM1 pKa = 7.35RR2 pKa = 11.84TACPTSFVGMWVPAVHH18 pKa = 7.25ADD20 pKa = 3.68CPHH23 pKa = 5.93NQVASLLKK31 pKa = 9.42RR32 pKa = 11.84TLGPVPDD39 pKa = 4.66PVHH42 pKa = 6.62EE43 pKa = 4.23PVGPTVKK50 pKa = 10.46AVFSRR55 pKa = 11.84LRR57 pKa = 11.84ALAARR62 pKa = 11.84SGVSRR67 pKa = 11.84WSLRR71 pKa = 11.84RR72 pKa = 11.84TAEE75 pKa = 4.06SYY77 pKa = 10.54KK78 pKa = 10.22GALGRR83 pKa = 11.84RR84 pKa = 11.84YY85 pKa = 9.19MEE87 pKa = 4.26AEE89 pKa = 3.45RR90 pKa = 11.84SLRR93 pKa = 11.84VDD95 pKa = 3.46GPVNSGDD102 pKa = 3.3TYY104 pKa = 11.36LRR106 pKa = 11.84PFLKK110 pKa = 10.59AEE112 pKa = 4.14KK113 pKa = 9.99VKK115 pKa = 11.01VMDD118 pKa = 4.0KK119 pKa = 10.3QSKK122 pKa = 9.27PRR124 pKa = 11.84MIFPRR129 pKa = 11.84SPRR132 pKa = 11.84YY133 pKa = 9.31NLSIASRR140 pKa = 11.84LKK142 pKa = 10.7PFEE145 pKa = 3.71HH146 pKa = 6.45WVWGRR151 pKa = 11.84LTTQVVLGTGVGRR164 pKa = 11.84VVAKK168 pKa = 10.32GLNPRR173 pKa = 11.84QRR175 pKa = 11.84ANLIVRR181 pKa = 11.84KK182 pKa = 9.49FRR184 pKa = 11.84ALDD187 pKa = 3.53DD188 pKa = 4.25CVCVEE193 pKa = 4.11VDD195 pKa = 3.6GKK197 pKa = 10.99AFEE200 pKa = 4.12AHH202 pKa = 5.87VGEE205 pKa = 4.52DD206 pKa = 3.39QLLEE210 pKa = 3.85EE211 pKa = 4.66HH212 pKa = 6.39GVYY215 pKa = 9.77EE216 pKa = 4.76AAFPGDD222 pKa = 3.43WEE224 pKa = 4.25LQRR227 pKa = 11.84LLRR230 pKa = 11.84VQLSLRR236 pKa = 11.84GKK238 pKa = 9.06LPCGAEE244 pKa = 4.05FTRR247 pKa = 11.84PGGRR251 pKa = 11.84ASGDD255 pKa = 3.49FNTGMGNSLIMLAVVGAVMKK275 pKa = 10.12EE276 pKa = 3.9LAPGAFDD283 pKa = 5.68LLVDD287 pKa = 4.48GDD289 pKa = 3.96NALVFMPRR297 pKa = 11.84GLLGLVRR304 pKa = 11.84ANFEE308 pKa = 4.03RR309 pKa = 11.84RR310 pKa = 11.84VLEE313 pKa = 4.06VSGHH317 pKa = 6.02EE318 pKa = 4.08IQLEE322 pKa = 4.58SPCSVIEE329 pKa = 4.24EE330 pKa = 4.08IRR332 pKa = 11.84FGQSAPINLGGSRR345 pKa = 11.84GWTMVRR351 pKa = 11.84DD352 pKa = 3.74PRR354 pKa = 11.84KK355 pKa = 10.33VISQALSSHH364 pKa = 5.72RR365 pKa = 11.84WLRR368 pKa = 11.84EE369 pKa = 3.41PTFASEE375 pKa = 4.74WIRR378 pKa = 11.84GVAACEE384 pKa = 3.99LSLSRR389 pKa = 11.84GVPVLQAWSASLQRR403 pKa = 11.84VYY405 pKa = 10.92GGPGGVRR412 pKa = 11.84EE413 pKa = 4.76HH414 pKa = 6.78PHH416 pKa = 5.64TDD418 pKa = 3.06YY419 pKa = 10.85FYY421 pKa = 10.59QGAWFADD428 pKa = 3.65VSQSVEE434 pKa = 3.86VTQQARR440 pKa = 11.84ISFWAAFGVAPDD452 pKa = 3.83EE453 pKa = 4.06QLALEE458 pKa = 4.81GGAFEE463 pKa = 5.13PSSEE467 pKa = 4.06DD468 pKa = 3.33YY469 pKa = 10.99EE470 pKa = 4.49RR471 pKa = 11.84VVLEE475 pKa = 4.01SFQWRR480 pKa = 11.84DD481 pKa = 3.42MPPGVAEE488 pKa = 4.41PLADD492 pKa = 3.67SLL494 pKa = 4.89
Molecular weight: 54.58 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7RYK7|A0A0A7RYK7_9VIRU RNA-dependent RNA polymerase OS=Magnaporthe oryzae RNA virus OX=1580607 GN=RdRp PE=4 SV=1
MM1 pKa = 7.71AFYY4 pKa = 10.71FDD6 pKa = 4.05PTYY9 pKa = 10.8HH10 pKa = 7.37ALGLNADD17 pKa = 3.44RR18 pKa = 11.84MMLGEE23 pKa = 4.32ALFVVKK29 pKa = 10.55VAGCFAAPVLISGVATVLRR48 pKa = 11.84GIFAVPPPGSWSPRR62 pKa = 11.84GFLTDD67 pKa = 3.18IGRR70 pKa = 11.84RR71 pKa = 11.84FATPEE76 pKa = 3.89TDD78 pKa = 3.41RR79 pKa = 11.84SGCGAIAPSSAVGITRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84ARR101 pKa = 11.84WVRR104 pKa = 11.84ALQDD108 pKa = 3.48MLGGPDD114 pKa = 3.41GCVGRR119 pKa = 11.84LFRR122 pKa = 11.84GRR124 pKa = 11.84WTPDD128 pKa = 2.85LPSSKK133 pKa = 9.66RR134 pKa = 11.84TDD136 pKa = 3.27VQNYY140 pKa = 9.3LLASIEE146 pKa = 4.16NGVKK150 pKa = 10.03ILGGGSEE157 pKa = 4.4KK158 pKa = 10.59VAPDD162 pKa = 3.6RR163 pKa = 11.84PSLDD167 pKa = 2.7GLYY170 pKa = 10.32FVVEE174 pKa = 3.9TSEE177 pKa = 4.5RR178 pKa = 11.84SVEE181 pKa = 4.18LVFPALLGRR190 pKa = 11.84LRR192 pKa = 11.84QYY194 pKa = 11.49ALWRR198 pKa = 11.84EE199 pKa = 4.04RR200 pKa = 11.84DD201 pKa = 3.71DD202 pKa = 3.9TLLGALRR209 pKa = 11.84TRR211 pKa = 11.84AVDD214 pKa = 3.1WCRR217 pKa = 11.84AVALRR222 pKa = 11.84PHH224 pKa = 5.85VADD227 pKa = 4.59LAVASCVSIAFEE239 pKa = 4.15PSLHH243 pKa = 6.02EE244 pKa = 4.16RR245 pKa = 11.84STRR248 pKa = 11.84PRR250 pKa = 11.84VDD252 pKa = 2.64RR253 pKa = 11.84HH254 pKa = 4.9MLRR257 pKa = 11.84SPGRR261 pKa = 11.84LSIYY265 pKa = 10.73
MM1 pKa = 7.71AFYY4 pKa = 10.71FDD6 pKa = 4.05PTYY9 pKa = 10.8HH10 pKa = 7.37ALGLNADD17 pKa = 3.44RR18 pKa = 11.84MMLGEE23 pKa = 4.32ALFVVKK29 pKa = 10.55VAGCFAAPVLISGVATVLRR48 pKa = 11.84GIFAVPPPGSWSPRR62 pKa = 11.84GFLTDD67 pKa = 3.18IGRR70 pKa = 11.84RR71 pKa = 11.84FATPEE76 pKa = 3.89TDD78 pKa = 3.41RR79 pKa = 11.84SGCGAIAPSSAVGITRR95 pKa = 11.84RR96 pKa = 11.84RR97 pKa = 11.84RR98 pKa = 11.84RR99 pKa = 11.84ARR101 pKa = 11.84WVRR104 pKa = 11.84ALQDD108 pKa = 3.48MLGGPDD114 pKa = 3.41GCVGRR119 pKa = 11.84LFRR122 pKa = 11.84GRR124 pKa = 11.84WTPDD128 pKa = 2.85LPSSKK133 pKa = 9.66RR134 pKa = 11.84TDD136 pKa = 3.27VQNYY140 pKa = 9.3LLASIEE146 pKa = 4.16NGVKK150 pKa = 10.03ILGGGSEE157 pKa = 4.4KK158 pKa = 10.59VAPDD162 pKa = 3.6RR163 pKa = 11.84PSLDD167 pKa = 2.7GLYY170 pKa = 10.32FVVEE174 pKa = 3.9TSEE177 pKa = 4.5RR178 pKa = 11.84SVEE181 pKa = 4.18LVFPALLGRR190 pKa = 11.84LRR192 pKa = 11.84QYY194 pKa = 11.49ALWRR198 pKa = 11.84EE199 pKa = 4.04RR200 pKa = 11.84DD201 pKa = 3.71DD202 pKa = 3.9TLLGALRR209 pKa = 11.84TRR211 pKa = 11.84AVDD214 pKa = 3.1WCRR217 pKa = 11.84AVALRR222 pKa = 11.84PHH224 pKa = 5.85VADD227 pKa = 4.59LAVASCVSIAFEE239 pKa = 4.15PSLHH243 pKa = 6.02EE244 pKa = 4.16RR245 pKa = 11.84STRR248 pKa = 11.84PRR250 pKa = 11.84VDD252 pKa = 2.64RR253 pKa = 11.84HH254 pKa = 4.9MLRR257 pKa = 11.84SPGRR261 pKa = 11.84LSIYY265 pKa = 10.73
Molecular weight: 29.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
759 |
265 |
494 |
379.5 |
41.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.486 ± 0.383 | 1.581 ± 0.167 |
4.743 ± 0.5 | 5.665 ± 1.03 |
4.216 ± 0.035 | 9.091 ± 0.019 |
1.845 ± 0.183 | 2.635 ± 0.415 |
2.503 ± 0.541 | 10.145 ± 0.435 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.108 ± 0.12 | 1.713 ± 0.316 |
6.588 ± 0.112 | 2.767 ± 0.89 |
10.277 ± 1.185 | 7.246 ± 0.042 |
3.557 ± 0.529 | 9.618 ± 0.511 |
2.24 ± 0.192 | 1.976 ± 0.157 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
