
Bacteroides sp. CAG:144
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; environmental samples
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
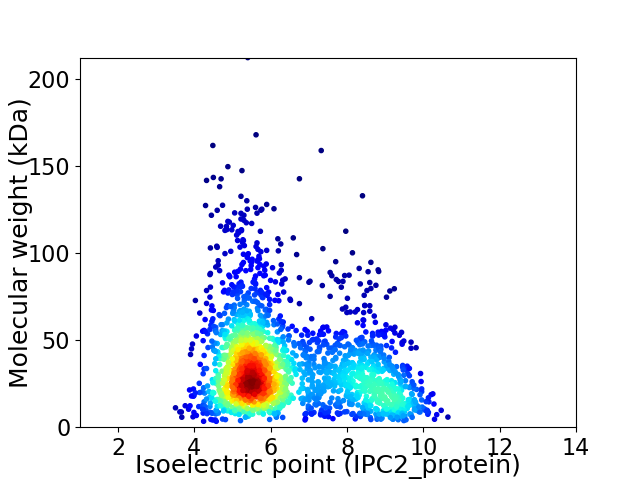
Virtual 2D-PAGE plot for 2103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5YAR8|R5YAR8_9BACE PadR domain-containing protein OS=Bacteroides sp. CAG:144 OX=1262736 GN=BN496_01736 PE=4 SV=1
MM1 pKa = 7.44KK2 pKa = 10.39KK3 pKa = 10.32KK4 pKa = 10.65LFLAMSLAMGLFVATSCSDD23 pKa = 3.56DD24 pKa = 5.24DD25 pKa = 4.55NDD27 pKa = 4.95GGNGGNPTINSNYY40 pKa = 9.85DD41 pKa = 3.32LAYY44 pKa = 10.2SAEE47 pKa = 4.16NATAWGNYY55 pKa = 4.86MVRR58 pKa = 11.84VAEE61 pKa = 4.55LLRR64 pKa = 11.84DD65 pKa = 3.94DD66 pKa = 5.38AATLYY71 pKa = 11.14ADD73 pKa = 3.95WNSSYY78 pKa = 11.61NGGASYY84 pKa = 11.51ASIFKK89 pKa = 10.21SHH91 pKa = 6.69NNDD94 pKa = 3.01TYY96 pKa = 11.59GSALSCIEE104 pKa = 4.55EE105 pKa = 4.55IIDD108 pKa = 3.9GCVTIADD115 pKa = 3.71EE116 pKa = 5.03VGDD119 pKa = 4.07NKK121 pKa = 10.99LGDD124 pKa = 4.1PYY126 pKa = 11.38DD127 pKa = 5.28LYY129 pKa = 10.91MQGNTTEE136 pKa = 3.87ALYY139 pKa = 10.53AVEE142 pKa = 5.54SWYY145 pKa = 10.67SWHH148 pKa = 7.26SRR150 pKa = 11.84DD151 pKa = 5.65DD152 pKa = 3.48YY153 pKa = 11.73TNNIWSIRR161 pKa = 11.84NAYY164 pKa = 9.77FGSLDD169 pKa = 3.41STVNANSISALVASVNTSLDD189 pKa = 3.61EE190 pKa = 4.09QMKK193 pKa = 8.85TAILNAAAAIQAIPQPLRR211 pKa = 11.84NNINSNEE218 pKa = 4.04TLEE221 pKa = 4.88AMEE224 pKa = 4.68ACADD228 pKa = 4.17LSAALSTLKK237 pKa = 10.61KK238 pKa = 10.47YY239 pKa = 10.48IQDD242 pKa = 3.81NLSDD246 pKa = 3.63NAALEE251 pKa = 4.84GIIDD255 pKa = 3.99TYY257 pKa = 11.15VDD259 pKa = 4.39DD260 pKa = 5.64VILPTYY266 pKa = 10.51QSLKK270 pKa = 10.2EE271 pKa = 4.23KK272 pKa = 11.06NSDD275 pKa = 3.39LYY277 pKa = 11.33DD278 pKa = 3.5AVVAFRR284 pKa = 11.84SNPSNAAFEE293 pKa = 4.63TACHH297 pKa = 5.43AWIEE301 pKa = 4.3ARR303 pKa = 11.84EE304 pKa = 3.97PWEE307 pKa = 3.89KK308 pKa = 11.11SEE310 pKa = 4.78AFLFGPVDD318 pKa = 4.4VEE320 pKa = 4.35GLDD323 pKa = 3.95PNMDD327 pKa = 3.55SWPLDD332 pKa = 2.87VDD334 pKa = 4.66AIVQILTTGNFGALDD349 pKa = 3.59WDD351 pKa = 4.41DD352 pKa = 5.12DD353 pKa = 4.57SEE355 pKa = 6.09AEE357 pKa = 3.93AAQSVRR363 pKa = 11.84GFHH366 pKa = 5.46TLEE369 pKa = 3.97FLLFQNGAPRR379 pKa = 11.84TIEE382 pKa = 3.73
MM1 pKa = 7.44KK2 pKa = 10.39KK3 pKa = 10.32KK4 pKa = 10.65LFLAMSLAMGLFVATSCSDD23 pKa = 3.56DD24 pKa = 5.24DD25 pKa = 4.55NDD27 pKa = 4.95GGNGGNPTINSNYY40 pKa = 9.85DD41 pKa = 3.32LAYY44 pKa = 10.2SAEE47 pKa = 4.16NATAWGNYY55 pKa = 4.86MVRR58 pKa = 11.84VAEE61 pKa = 4.55LLRR64 pKa = 11.84DD65 pKa = 3.94DD66 pKa = 5.38AATLYY71 pKa = 11.14ADD73 pKa = 3.95WNSSYY78 pKa = 11.61NGGASYY84 pKa = 11.51ASIFKK89 pKa = 10.21SHH91 pKa = 6.69NNDD94 pKa = 3.01TYY96 pKa = 11.59GSALSCIEE104 pKa = 4.55EE105 pKa = 4.55IIDD108 pKa = 3.9GCVTIADD115 pKa = 3.71EE116 pKa = 5.03VGDD119 pKa = 4.07NKK121 pKa = 10.99LGDD124 pKa = 4.1PYY126 pKa = 11.38DD127 pKa = 5.28LYY129 pKa = 10.91MQGNTTEE136 pKa = 3.87ALYY139 pKa = 10.53AVEE142 pKa = 5.54SWYY145 pKa = 10.67SWHH148 pKa = 7.26SRR150 pKa = 11.84DD151 pKa = 5.65DD152 pKa = 3.48YY153 pKa = 11.73TNNIWSIRR161 pKa = 11.84NAYY164 pKa = 9.77FGSLDD169 pKa = 3.41STVNANSISALVASVNTSLDD189 pKa = 3.61EE190 pKa = 4.09QMKK193 pKa = 8.85TAILNAAAAIQAIPQPLRR211 pKa = 11.84NNINSNEE218 pKa = 4.04TLEE221 pKa = 4.88AMEE224 pKa = 4.68ACADD228 pKa = 4.17LSAALSTLKK237 pKa = 10.61KK238 pKa = 10.47YY239 pKa = 10.48IQDD242 pKa = 3.81NLSDD246 pKa = 3.63NAALEE251 pKa = 4.84GIIDD255 pKa = 3.99TYY257 pKa = 11.15VDD259 pKa = 4.39DD260 pKa = 5.64VILPTYY266 pKa = 10.51QSLKK270 pKa = 10.2EE271 pKa = 4.23KK272 pKa = 11.06NSDD275 pKa = 3.39LYY277 pKa = 11.33DD278 pKa = 3.5AVVAFRR284 pKa = 11.84SNPSNAAFEE293 pKa = 4.63TACHH297 pKa = 5.43AWIEE301 pKa = 4.3ARR303 pKa = 11.84EE304 pKa = 3.97PWEE307 pKa = 3.89KK308 pKa = 11.11SEE310 pKa = 4.78AFLFGPVDD318 pKa = 4.4VEE320 pKa = 4.35GLDD323 pKa = 3.95PNMDD327 pKa = 3.55SWPLDD332 pKa = 2.87VDD334 pKa = 4.66AIVQILTTGNFGALDD349 pKa = 3.59WDD351 pKa = 4.41DD352 pKa = 5.12DD353 pKa = 4.57SEE355 pKa = 6.09AEE357 pKa = 3.93AAQSVRR363 pKa = 11.84GFHH366 pKa = 5.46TLEE369 pKa = 3.97FLLFQNGAPRR379 pKa = 11.84TIEE382 pKa = 3.73
Molecular weight: 41.74 kDa
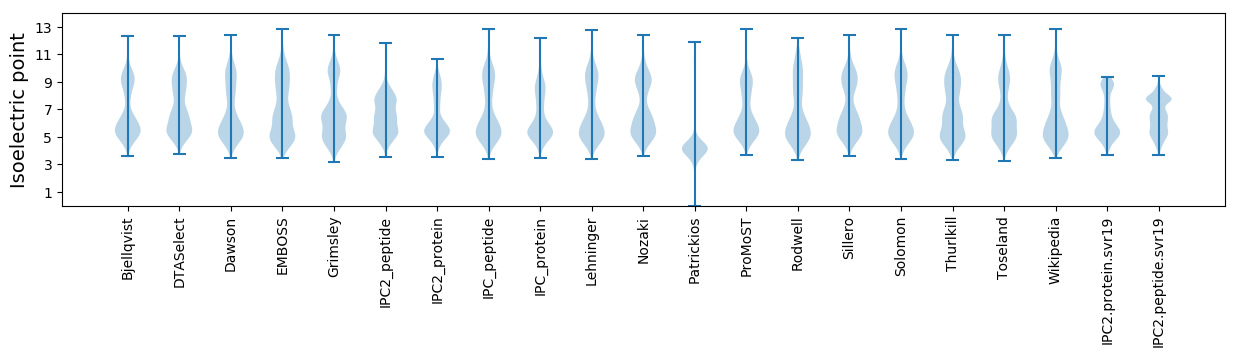
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5Y9F7|R5Y9F7_9BACE SAF domain-containing protein OS=Bacteroides sp. CAG:144 OX=1262736 GN=BN496_01316 PE=3 SV=1
MM1 pKa = 6.22MRR3 pKa = 11.84AWARR7 pKa = 11.84NVSSPSFRR15 pKa = 11.84LMEE18 pKa = 3.85LTIPFPCIHH27 pKa = 6.86FSPARR32 pKa = 11.84MTGHH36 pKa = 6.5FEE38 pKa = 3.55EE39 pKa = 5.49SIIIGTRR46 pKa = 11.84QISGSEE52 pKa = 3.55AMRR55 pKa = 11.84FRR57 pKa = 11.84NVTISFCASSMASSILMSMICAPSSTCLRR86 pKa = 11.84AIPRR90 pKa = 11.84ASSYY94 pKa = 10.44FFSEE98 pKa = 4.19ISRR101 pKa = 11.84KK102 pKa = 7.63NLRR105 pKa = 11.84EE106 pKa = 3.69PATLHH111 pKa = 6.88RR112 pKa = 11.84SPTLIKK118 pKa = 10.26LFSGMIFNSSKK129 pKa = 10.18PDD131 pKa = 3.41KK132 pKa = 10.75QSTFSLICGRR142 pKa = 4.19
MM1 pKa = 6.22MRR3 pKa = 11.84AWARR7 pKa = 11.84NVSSPSFRR15 pKa = 11.84LMEE18 pKa = 3.85LTIPFPCIHH27 pKa = 6.86FSPARR32 pKa = 11.84MTGHH36 pKa = 6.5FEE38 pKa = 3.55EE39 pKa = 5.49SIIIGTRR46 pKa = 11.84QISGSEE52 pKa = 3.55AMRR55 pKa = 11.84FRR57 pKa = 11.84NVTISFCASSMASSILMSMICAPSSTCLRR86 pKa = 11.84AIPRR90 pKa = 11.84ASSYY94 pKa = 10.44FFSEE98 pKa = 4.19ISRR101 pKa = 11.84KK102 pKa = 7.63NLRR105 pKa = 11.84EE106 pKa = 3.69PATLHH111 pKa = 6.88RR112 pKa = 11.84SPTLIKK118 pKa = 10.26LFSGMIFNSSKK129 pKa = 10.18PDD131 pKa = 3.41KK132 pKa = 10.75QSTFSLICGRR142 pKa = 4.19
Molecular weight: 15.9 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691520 |
29 |
1880 |
328.8 |
37.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.257 ± 0.053 | 1.336 ± 0.021 |
5.65 ± 0.043 | 6.387 ± 0.047 |
4.669 ± 0.039 | 6.954 ± 0.048 |
1.945 ± 0.024 | 7.324 ± 0.052 |
5.738 ± 0.044 | 9.094 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.537 ± 0.025 | 4.587 ± 0.037 |
3.909 ± 0.025 | 3.168 ± 0.029 |
5.292 ± 0.04 | 6.316 ± 0.041 |
5.631 ± 0.047 | 6.621 ± 0.046 |
1.147 ± 0.02 | 4.434 ± 0.041 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
