
Pseudomonas mendocina (strain ymp)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas aeruginosa group; Pseudomonas mendocina
Average proteome isoelectric point is 6.32
Get precalculated fractions of proteins
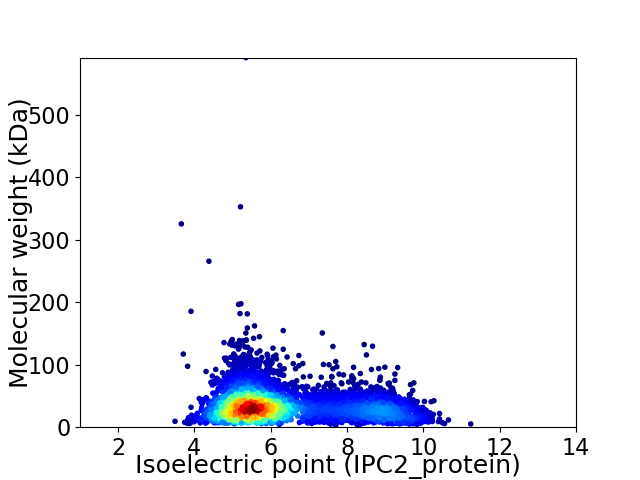
Virtual 2D-PAGE plot for 4563 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
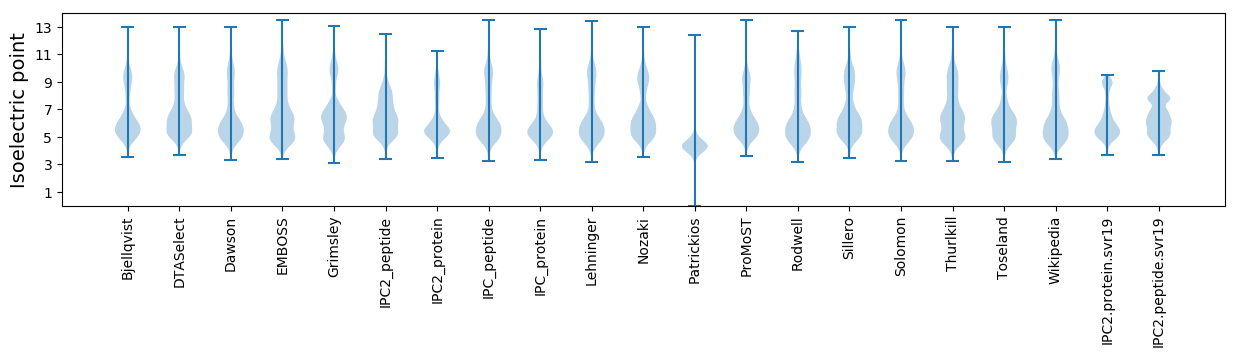
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4XN66|A4XN66_PSEMY Uncharacterized protein OS=Pseudomonas mendocina (strain ymp) OX=399739 GN=Pmen_0007 PE=4 SV=1
MM1 pKa = 7.62TNYY4 pKa = 9.92QQITPFSYY12 pKa = 10.15IPLSISATVNTLLYY26 pKa = 7.54GTKK29 pKa = 9.41WGGGLGFPVTLTYY42 pKa = 11.12SFANPGVSQYY52 pKa = 10.18STDD55 pKa = 3.34YY56 pKa = 11.66DD57 pKa = 3.96DD58 pKa = 6.2FVEE61 pKa = 4.6YY62 pKa = 7.89WTGSNSSNFIYY73 pKa = 10.54SLSSNQEE80 pKa = 3.76SAVRR84 pKa = 11.84SALAQWSSVANLSFTEE100 pKa = 4.21VSDD103 pKa = 3.86TAGGTVGDD111 pKa = 4.48LRR113 pKa = 11.84FGGYY117 pKa = 10.42KK118 pKa = 10.58LIDD121 pKa = 3.26WDD123 pKa = 3.84TAAFAFLPGASPLAGDD139 pKa = 3.52VWIGPVTNEE148 pKa = 4.0VNPSGGSYY156 pKa = 10.7DD157 pKa = 3.84YY158 pKa = 10.89LTFLHH163 pKa = 6.95EE164 pKa = 4.63IGHH167 pKa = 6.36ALGLKK172 pKa = 10.22HH173 pKa = 6.57PFEE176 pKa = 5.92ASGSSNYY183 pKa = 8.99VLRR186 pKa = 11.84GAYY189 pKa = 10.06DD190 pKa = 3.49DD191 pKa = 3.74VRR193 pKa = 11.84YY194 pKa = 7.89TVMSYY199 pKa = 11.34NDD201 pKa = 3.8TYY203 pKa = 11.83DD204 pKa = 3.87FLPTGPMLYY213 pKa = 10.22DD214 pKa = 3.75IAAMQYY220 pKa = 10.05IYY222 pKa = 10.93GSNDD226 pKa = 2.33TWQTGDD232 pKa = 4.69NVYY235 pKa = 10.65SWDD238 pKa = 3.31ANARR242 pKa = 11.84IFEE245 pKa = 4.74TIWDD249 pKa = 3.72AGGNDD254 pKa = 4.31TIDD257 pKa = 4.51ASNQSAGVVINLNSGEE273 pKa = 4.12FSSIGQSIYY282 pKa = 11.16NYY284 pKa = 9.24ATGAGRR290 pKa = 11.84NNFLTIAYY298 pKa = 9.73GSVIEE303 pKa = 4.19NAIGSDD309 pKa = 3.8YY310 pKa = 11.28KK311 pKa = 10.78DD312 pKa = 3.37TLIGNSADD320 pKa = 3.53NVLDD324 pKa = 3.9GGKK327 pKa = 10.65GEE329 pKa = 4.15DD330 pKa = 3.8VLRR333 pKa = 11.84GGAGNDD339 pKa = 3.26TYY341 pKa = 11.7YY342 pKa = 11.33VDD344 pKa = 5.47LPTGSDD350 pKa = 3.33FYY352 pKa = 11.84DD353 pKa = 3.29RR354 pKa = 11.84VTEE357 pKa = 4.11YY358 pKa = 11.04AGEE361 pKa = 4.71GYY363 pKa = 10.78DD364 pKa = 3.86KK365 pKa = 11.21VVLRR369 pKa = 11.84GTSTGFFGRR378 pKa = 11.84SFALANNVEE387 pKa = 4.49EE388 pKa = 4.38VDD390 pKa = 3.62ASQTGAVALTLLGNNSANRR409 pKa = 11.84IIGNAGNNLLNGGAGADD426 pKa = 3.74TMIGGAGNDD435 pKa = 3.83TYY437 pKa = 11.68VVDD440 pKa = 3.83HH441 pKa = 6.98LGDD444 pKa = 3.65EE445 pKa = 4.78VIEE448 pKa = 4.14QVGEE452 pKa = 4.46GIDD455 pKa = 3.71LVQVAIATAGGTYY468 pKa = 9.85TLGDD472 pKa = 3.71HH473 pKa = 6.67VEE475 pKa = 4.25NATLTNSVSFNLVGNTLSNTLVGNAANNLLDD506 pKa = 4.23GGAGADD512 pKa = 3.8VMNGGAGNDD521 pKa = 3.69TYY523 pKa = 11.69VVDD526 pKa = 6.13DD527 pKa = 4.92IGDD530 pKa = 3.77QIIDD534 pKa = 3.52SAGIDD539 pKa = 3.43TVLASISYY547 pKa = 8.81TLGNDD552 pKa = 3.85LEE554 pKa = 4.29NLSLLGSEE562 pKa = 4.71ALNGTGNGLANVITGNDD579 pKa = 3.61GSNVLDD585 pKa = 3.98GGGGVDD591 pKa = 3.54TLRR594 pKa = 11.84GGKK597 pKa = 10.24GDD599 pKa = 3.51DD600 pKa = 3.61TYY602 pKa = 11.35IVDD605 pKa = 4.17LTARR609 pKa = 11.84NTLEE613 pKa = 5.05DD614 pKa = 3.66SVLEE618 pKa = 3.95NLYY621 pKa = 10.4EE622 pKa = 4.51GNDD625 pKa = 3.96TIQLRR630 pKa = 11.84GGNVALATVTAIALANNVEE649 pKa = 4.21NLDD652 pKa = 3.7ASATGTTRR660 pKa = 11.84LNLTGNVLNNILIGNAADD678 pKa = 3.54NLLNGGAGADD688 pKa = 3.74RR689 pKa = 11.84LIGGDD694 pKa = 3.67GNDD697 pKa = 3.49TYY699 pKa = 11.8VVDD702 pKa = 3.8NLGDD706 pKa = 3.79EE707 pKa = 4.88VIEE710 pKa = 4.16QAGEE714 pKa = 4.16GTDD717 pKa = 3.66LVQVAIATAGGTYY730 pKa = 9.79TLGEE734 pKa = 4.22HH735 pKa = 6.25VEE737 pKa = 4.25NATLTNKK744 pKa = 9.53VAFNLTGNALSNTLVGNAANNVLDD768 pKa = 4.47GGLGADD774 pKa = 4.09VMNGGAGNDD783 pKa = 3.66TYY785 pKa = 11.14IVDD788 pKa = 4.4DD789 pKa = 4.34VGDD792 pKa = 4.15TIIDD796 pKa = 3.55SAGIDD801 pKa = 3.42TVRR804 pKa = 11.84ASIAYY809 pKa = 7.77TLGKK813 pKa = 10.42GLEE816 pKa = 4.1NLVLTGSHH824 pKa = 6.01AVSGTGNALANILDD838 pKa = 4.3GSQNNAANEE847 pKa = 3.72LRR849 pKa = 11.84GLGGNDD855 pKa = 3.39TYY857 pKa = 11.18IVGAGDD863 pKa = 3.74TVIEE867 pKa = 4.24EE868 pKa = 4.4ANGGIDD874 pKa = 4.05LVQAYY879 pKa = 9.61VSYY882 pKa = 9.93TLGDD886 pKa = 3.59HH887 pKa = 6.76VEE889 pKa = 4.06NLSLLGSEE897 pKa = 4.71ALNGTGNGLANVLTGNDD914 pKa = 3.65GSNVLDD920 pKa = 3.98GGGGVDD926 pKa = 3.54TLRR929 pKa = 11.84GGKK932 pKa = 10.24GDD934 pKa = 3.51DD935 pKa = 3.61TYY937 pKa = 11.37IVDD940 pKa = 4.97LMANNALEE948 pKa = 4.2DD949 pKa = 3.56RR950 pKa = 11.84VVEE953 pKa = 4.11NANEE957 pKa = 4.68GIDD960 pKa = 3.86TIVLRR965 pKa = 11.84GGSALPKK972 pKa = 10.15AATIVLANNVEE983 pKa = 4.07NLDD986 pKa = 3.64ASGTGSIALNLTGNAANNLLIGNQGNNTLNGGDD1019 pKa = 4.38GNDD1022 pKa = 3.64TLIGMGGFDD1031 pKa = 3.91TLTGGRR1037 pKa = 11.84GADD1040 pKa = 3.26TFRR1043 pKa = 11.84FALTWLGMNSGPDD1056 pKa = 3.32AVITDD1061 pKa = 3.82FRR1063 pKa = 11.84RR1064 pKa = 11.84LDD1066 pKa = 3.37GDD1068 pKa = 4.15KK1069 pKa = 10.68IQLQGEE1075 pKa = 4.1DD1076 pKa = 3.33AEE1078 pKa = 4.43RR1079 pKa = 11.84FSFVGRR1085 pKa = 11.84TFDD1088 pKa = 3.11TGATGSHH1095 pKa = 5.55QLRR1098 pKa = 11.84FDD1100 pKa = 3.54AGTLYY1105 pKa = 11.11GSVDD1109 pKa = 3.36SGMSNAFEE1117 pKa = 3.85IRR1119 pKa = 11.84LLGVSEE1125 pKa = 4.57LRR1127 pKa = 11.84QDD1129 pKa = 3.95DD1130 pKa = 4.16FLFGG1134 pKa = 3.75
MM1 pKa = 7.62TNYY4 pKa = 9.92QQITPFSYY12 pKa = 10.15IPLSISATVNTLLYY26 pKa = 7.54GTKK29 pKa = 9.41WGGGLGFPVTLTYY42 pKa = 11.12SFANPGVSQYY52 pKa = 10.18STDD55 pKa = 3.34YY56 pKa = 11.66DD57 pKa = 3.96DD58 pKa = 6.2FVEE61 pKa = 4.6YY62 pKa = 7.89WTGSNSSNFIYY73 pKa = 10.54SLSSNQEE80 pKa = 3.76SAVRR84 pKa = 11.84SALAQWSSVANLSFTEE100 pKa = 4.21VSDD103 pKa = 3.86TAGGTVGDD111 pKa = 4.48LRR113 pKa = 11.84FGGYY117 pKa = 10.42KK118 pKa = 10.58LIDD121 pKa = 3.26WDD123 pKa = 3.84TAAFAFLPGASPLAGDD139 pKa = 3.52VWIGPVTNEE148 pKa = 4.0VNPSGGSYY156 pKa = 10.7DD157 pKa = 3.84YY158 pKa = 10.89LTFLHH163 pKa = 6.95EE164 pKa = 4.63IGHH167 pKa = 6.36ALGLKK172 pKa = 10.22HH173 pKa = 6.57PFEE176 pKa = 5.92ASGSSNYY183 pKa = 8.99VLRR186 pKa = 11.84GAYY189 pKa = 10.06DD190 pKa = 3.49DD191 pKa = 3.74VRR193 pKa = 11.84YY194 pKa = 7.89TVMSYY199 pKa = 11.34NDD201 pKa = 3.8TYY203 pKa = 11.83DD204 pKa = 3.87FLPTGPMLYY213 pKa = 10.22DD214 pKa = 3.75IAAMQYY220 pKa = 10.05IYY222 pKa = 10.93GSNDD226 pKa = 2.33TWQTGDD232 pKa = 4.69NVYY235 pKa = 10.65SWDD238 pKa = 3.31ANARR242 pKa = 11.84IFEE245 pKa = 4.74TIWDD249 pKa = 3.72AGGNDD254 pKa = 4.31TIDD257 pKa = 4.51ASNQSAGVVINLNSGEE273 pKa = 4.12FSSIGQSIYY282 pKa = 11.16NYY284 pKa = 9.24ATGAGRR290 pKa = 11.84NNFLTIAYY298 pKa = 9.73GSVIEE303 pKa = 4.19NAIGSDD309 pKa = 3.8YY310 pKa = 11.28KK311 pKa = 10.78DD312 pKa = 3.37TLIGNSADD320 pKa = 3.53NVLDD324 pKa = 3.9GGKK327 pKa = 10.65GEE329 pKa = 4.15DD330 pKa = 3.8VLRR333 pKa = 11.84GGAGNDD339 pKa = 3.26TYY341 pKa = 11.7YY342 pKa = 11.33VDD344 pKa = 5.47LPTGSDD350 pKa = 3.33FYY352 pKa = 11.84DD353 pKa = 3.29RR354 pKa = 11.84VTEE357 pKa = 4.11YY358 pKa = 11.04AGEE361 pKa = 4.71GYY363 pKa = 10.78DD364 pKa = 3.86KK365 pKa = 11.21VVLRR369 pKa = 11.84GTSTGFFGRR378 pKa = 11.84SFALANNVEE387 pKa = 4.49EE388 pKa = 4.38VDD390 pKa = 3.62ASQTGAVALTLLGNNSANRR409 pKa = 11.84IIGNAGNNLLNGGAGADD426 pKa = 3.74TMIGGAGNDD435 pKa = 3.83TYY437 pKa = 11.68VVDD440 pKa = 3.83HH441 pKa = 6.98LGDD444 pKa = 3.65EE445 pKa = 4.78VIEE448 pKa = 4.14QVGEE452 pKa = 4.46GIDD455 pKa = 3.71LVQVAIATAGGTYY468 pKa = 9.85TLGDD472 pKa = 3.71HH473 pKa = 6.67VEE475 pKa = 4.25NATLTNSVSFNLVGNTLSNTLVGNAANNLLDD506 pKa = 4.23GGAGADD512 pKa = 3.8VMNGGAGNDD521 pKa = 3.69TYY523 pKa = 11.69VVDD526 pKa = 6.13DD527 pKa = 4.92IGDD530 pKa = 3.77QIIDD534 pKa = 3.52SAGIDD539 pKa = 3.43TVLASISYY547 pKa = 8.81TLGNDD552 pKa = 3.85LEE554 pKa = 4.29NLSLLGSEE562 pKa = 4.71ALNGTGNGLANVITGNDD579 pKa = 3.61GSNVLDD585 pKa = 3.98GGGGVDD591 pKa = 3.54TLRR594 pKa = 11.84GGKK597 pKa = 10.24GDD599 pKa = 3.51DD600 pKa = 3.61TYY602 pKa = 11.35IVDD605 pKa = 4.17LTARR609 pKa = 11.84NTLEE613 pKa = 5.05DD614 pKa = 3.66SVLEE618 pKa = 3.95NLYY621 pKa = 10.4EE622 pKa = 4.51GNDD625 pKa = 3.96TIQLRR630 pKa = 11.84GGNVALATVTAIALANNVEE649 pKa = 4.21NLDD652 pKa = 3.7ASATGTTRR660 pKa = 11.84LNLTGNVLNNILIGNAADD678 pKa = 3.54NLLNGGAGADD688 pKa = 3.74RR689 pKa = 11.84LIGGDD694 pKa = 3.67GNDD697 pKa = 3.49TYY699 pKa = 11.8VVDD702 pKa = 3.8NLGDD706 pKa = 3.79EE707 pKa = 4.88VIEE710 pKa = 4.16QAGEE714 pKa = 4.16GTDD717 pKa = 3.66LVQVAIATAGGTYY730 pKa = 9.79TLGEE734 pKa = 4.22HH735 pKa = 6.25VEE737 pKa = 4.25NATLTNKK744 pKa = 9.53VAFNLTGNALSNTLVGNAANNVLDD768 pKa = 4.47GGLGADD774 pKa = 4.09VMNGGAGNDD783 pKa = 3.66TYY785 pKa = 11.14IVDD788 pKa = 4.4DD789 pKa = 4.34VGDD792 pKa = 4.15TIIDD796 pKa = 3.55SAGIDD801 pKa = 3.42TVRR804 pKa = 11.84ASIAYY809 pKa = 7.77TLGKK813 pKa = 10.42GLEE816 pKa = 4.1NLVLTGSHH824 pKa = 6.01AVSGTGNALANILDD838 pKa = 4.3GSQNNAANEE847 pKa = 3.72LRR849 pKa = 11.84GLGGNDD855 pKa = 3.39TYY857 pKa = 11.18IVGAGDD863 pKa = 3.74TVIEE867 pKa = 4.24EE868 pKa = 4.4ANGGIDD874 pKa = 4.05LVQAYY879 pKa = 9.61VSYY882 pKa = 9.93TLGDD886 pKa = 3.59HH887 pKa = 6.76VEE889 pKa = 4.06NLSLLGSEE897 pKa = 4.71ALNGTGNGLANVLTGNDD914 pKa = 3.65GSNVLDD920 pKa = 3.98GGGGVDD926 pKa = 3.54TLRR929 pKa = 11.84GGKK932 pKa = 10.24GDD934 pKa = 3.51DD935 pKa = 3.61TYY937 pKa = 11.37IVDD940 pKa = 4.97LMANNALEE948 pKa = 4.2DD949 pKa = 3.56RR950 pKa = 11.84VVEE953 pKa = 4.11NANEE957 pKa = 4.68GIDD960 pKa = 3.86TIVLRR965 pKa = 11.84GGSALPKK972 pKa = 10.15AATIVLANNVEE983 pKa = 4.07NLDD986 pKa = 3.64ASGTGSIALNLTGNAANNLLIGNQGNNTLNGGDD1019 pKa = 4.38GNDD1022 pKa = 3.64TLIGMGGFDD1031 pKa = 3.91TLTGGRR1037 pKa = 11.84GADD1040 pKa = 3.26TFRR1043 pKa = 11.84FALTWLGMNSGPDD1056 pKa = 3.32AVITDD1061 pKa = 3.82FRR1063 pKa = 11.84RR1064 pKa = 11.84LDD1066 pKa = 3.37GDD1068 pKa = 4.15KK1069 pKa = 10.68IQLQGEE1075 pKa = 4.1DD1076 pKa = 3.33AEE1078 pKa = 4.43RR1079 pKa = 11.84FSFVGRR1085 pKa = 11.84TFDD1088 pKa = 3.11TGATGSHH1095 pKa = 5.55QLRR1098 pKa = 11.84FDD1100 pKa = 3.54AGTLYY1105 pKa = 11.11GSVDD1109 pKa = 3.36SGMSNAFEE1117 pKa = 3.85IRR1119 pKa = 11.84LLGVSEE1125 pKa = 4.57LRR1127 pKa = 11.84QDD1129 pKa = 3.95DD1130 pKa = 4.16FLFGG1134 pKa = 3.75
Molecular weight: 117.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4XNA6|A4XNA6_PSEMY Type III secretion protein SpaR/YscT/HrcT OS=Pseudomonas mendocina (strain ymp) OX=399739 GN=Pmen_0048 PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTIKK11 pKa = 10.52RR12 pKa = 11.84ARR14 pKa = 11.84THH16 pKa = 5.94GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.16NGRR28 pKa = 11.84AVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.15 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1497472 |
23 |
5328 |
328.2 |
36.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.557 ± 0.05 | 1.007 ± 0.011 |
5.281 ± 0.03 | 6.101 ± 0.03 |
3.528 ± 0.021 | 8.033 ± 0.034 |
2.258 ± 0.019 | 4.507 ± 0.026 |
3.094 ± 0.028 | 12.493 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.211 ± 0.018 | 2.742 ± 0.024 |
4.894 ± 0.025 | 4.798 ± 0.032 |
6.955 ± 0.039 | 5.518 ± 0.027 |
4.285 ± 0.025 | 6.796 ± 0.03 |
1.455 ± 0.016 | 2.488 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
