
Mycoplasma bovoculi M165/69
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Tenericutes; Mollicutes; Mycoplasmatales; Mycoplasmataceae; Mycoplasma; Mycoplasma bovoculi
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
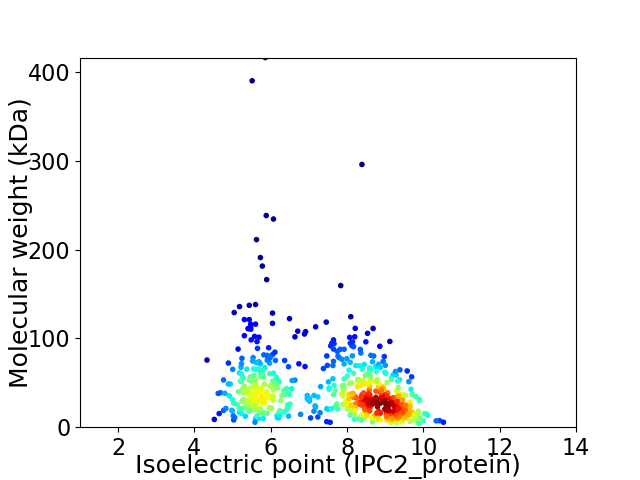
Virtual 2D-PAGE plot for 579 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W5USN2|W5USN2_9MOLU Guanylate kinase OS=Mycoplasma bovoculi M165/69 OX=743966 GN=gmk PE=3 SV=1
MM1 pKa = 7.66GKK3 pKa = 9.82NKK5 pKa = 9.71ILKK8 pKa = 9.34FYY10 pKa = 10.83SSEE13 pKa = 3.78LCYY16 pKa = 10.7EE17 pKa = 4.05NNGQRR22 pKa = 11.84VCQHH26 pKa = 5.52SLRR29 pKa = 11.84YY30 pKa = 9.47DD31 pKa = 3.19KK32 pKa = 10.3QTIGVFANFFDD43 pKa = 4.81LLVKK47 pKa = 10.42INSLNLVARR56 pKa = 11.84LWVRR60 pKa = 11.84QNKK63 pKa = 9.18KK64 pKa = 9.27FSLVVRR70 pKa = 11.84FPFKK74 pKa = 10.65RR75 pKa = 11.84SLDD78 pKa = 3.82SLNTADD84 pKa = 5.13IQMLLDD90 pKa = 4.9KK91 pKa = 10.71IADD94 pKa = 3.85PSSTGDD100 pKa = 3.48IIEE103 pKa = 3.99QLQYY107 pKa = 10.66TEE109 pKa = 4.49EE110 pKa = 4.27EE111 pKa = 4.3FVQVLEE117 pKa = 4.24EE118 pKa = 4.0QNNNKK123 pKa = 9.66DD124 pKa = 3.74LNISKK129 pKa = 8.94TINKK133 pKa = 9.3ILFDD137 pKa = 4.1VEE139 pKa = 4.99SDD141 pKa = 3.93DD142 pKa = 5.13EE143 pKa = 4.21PVEE146 pKa = 4.86CEE148 pKa = 3.5ICQEE152 pKa = 4.15TLRR155 pKa = 11.84EE156 pKa = 4.05DD157 pKa = 4.99EE158 pKa = 4.75IDD160 pKa = 4.01CGCDD164 pKa = 2.76SIEE167 pKa = 3.64IHH169 pKa = 6.51RR170 pKa = 11.84ADD172 pKa = 4.75FEE174 pKa = 4.88GFSCDD179 pKa = 5.02CPICLSDD186 pKa = 3.64KK187 pKa = 10.42HH188 pKa = 6.25AHH190 pKa = 5.71TYY192 pKa = 9.55EE193 pKa = 3.99EE194 pKa = 4.36PRR196 pKa = 11.84VEE198 pKa = 4.45EE199 pKa = 4.14EE200 pKa = 3.74PLYY203 pKa = 11.03LCEE206 pKa = 4.34CAKK209 pKa = 10.82CLGQEE214 pKa = 3.55VDD216 pKa = 3.56EE217 pKa = 4.53PLNIIEE223 pKa = 4.45EE224 pKa = 4.38TDD226 pKa = 3.99CKK228 pKa = 10.97CEE230 pKa = 4.07SCVKK234 pKa = 10.51GFDD237 pKa = 4.16DD238 pKa = 4.74IKK240 pKa = 10.81PIIIDD245 pKa = 3.6EE246 pKa = 4.56PTTQDD251 pKa = 4.29YY252 pKa = 8.65PCHH255 pKa = 6.0CQNNQQSNAQLIQTYY270 pKa = 9.55FDD272 pKa = 3.52EE273 pKa = 5.21SLYY276 pKa = 11.23SCNCVDD282 pKa = 4.61CASEE286 pKa = 4.01QKK288 pKa = 10.2QVEE291 pKa = 4.42EE292 pKa = 4.71FVDD295 pKa = 4.04EE296 pKa = 4.41PEE298 pKa = 4.35IKK300 pKa = 10.69VGTLLSNSCKK310 pKa = 10.27GVRR313 pKa = 11.84CPISKK318 pKa = 10.52EE319 pKa = 4.04EE320 pKa = 4.15TSDD323 pKa = 3.57EE324 pKa = 4.33PKK326 pKa = 10.49TEE328 pKa = 3.97VLEE331 pKa = 4.09PTIDD335 pKa = 3.66EE336 pKa = 4.49SKK338 pKa = 9.47TAYY341 pKa = 8.85EE342 pKa = 4.06LPYY345 pKa = 10.59EE346 pKa = 4.59CFCHH350 pKa = 6.67EE351 pKa = 4.55CQNPVPSNLDD361 pKa = 3.03YY362 pKa = 10.96FLWFKK367 pKa = 10.73QSQPKK372 pKa = 9.76QIDD375 pKa = 3.24YY376 pKa = 8.38TLPYY380 pKa = 9.96EE381 pKa = 4.63CQCHH385 pKa = 5.72LCSGQEE391 pKa = 3.79IEE393 pKa = 5.83LSLDD397 pKa = 3.5YY398 pKa = 10.84ILAEE402 pKa = 3.73TSVNKK407 pKa = 9.92EE408 pKa = 3.58QLEE411 pKa = 4.26EE412 pKa = 3.92VLIADD417 pKa = 4.12EE418 pKa = 5.3PIIVDD423 pKa = 3.44NFEE426 pKa = 4.4SKK428 pKa = 10.9SEE430 pKa = 3.88ILCNCDD436 pKa = 4.08HH437 pKa = 6.81NEE439 pKa = 3.92CDD441 pKa = 4.57EE442 pKa = 4.52PQLDD446 pKa = 4.22PIVNLDD452 pKa = 3.91HH453 pKa = 7.35LNAPLLAPTTTATNFEE469 pKa = 4.73CPACQHH475 pKa = 5.94VATCDD480 pKa = 3.26FCQKK484 pKa = 10.51YY485 pKa = 10.19KK486 pKa = 11.02LYY488 pKa = 10.39LLEE491 pKa = 6.12AIEE494 pKa = 5.5CPGCQKK500 pKa = 10.71LFADD504 pKa = 4.28RR505 pKa = 11.84GISIEE510 pKa = 4.32EE511 pKa = 3.92LLKK514 pKa = 10.95SYY516 pKa = 10.13FPHH519 pKa = 7.22ISYY522 pKa = 10.41DD523 pKa = 3.82GKK525 pKa = 10.71SHH527 pKa = 7.0GSQLCSLSTSEE538 pKa = 5.08QEE540 pKa = 4.72CPLAKK545 pKa = 10.6LKK547 pKa = 10.33DD548 pKa = 3.51QKK550 pKa = 11.11FIDD553 pKa = 3.63LHH555 pKa = 6.9DD556 pKa = 4.22EE557 pKa = 3.91NRR559 pKa = 11.84GIIYY563 pKa = 9.3TPEE566 pKa = 5.76DD567 pKa = 3.83GVSSGDD573 pKa = 3.47YY574 pKa = 11.11DD575 pKa = 3.68EE576 pKa = 6.01LLEE579 pKa = 4.83GNVNTKK585 pKa = 10.02RR586 pKa = 11.84GYY588 pKa = 10.36GGGGVGGPATGQGVGRR604 pKa = 11.84FAVDD608 pKa = 3.25DD609 pKa = 3.9TSAIIGAVSEE619 pKa = 4.25KK620 pKa = 10.67NKK622 pKa = 9.91PDD624 pKa = 3.5EE625 pKa = 4.78EE626 pKa = 4.39IVLVTTKK633 pKa = 9.54KK634 pKa = 8.88TGFWGIPAYY643 pKa = 9.81VLWFIVFALIAAIIIIVVMFFVFGSGLII671 pKa = 3.78
MM1 pKa = 7.66GKK3 pKa = 9.82NKK5 pKa = 9.71ILKK8 pKa = 9.34FYY10 pKa = 10.83SSEE13 pKa = 3.78LCYY16 pKa = 10.7EE17 pKa = 4.05NNGQRR22 pKa = 11.84VCQHH26 pKa = 5.52SLRR29 pKa = 11.84YY30 pKa = 9.47DD31 pKa = 3.19KK32 pKa = 10.3QTIGVFANFFDD43 pKa = 4.81LLVKK47 pKa = 10.42INSLNLVARR56 pKa = 11.84LWVRR60 pKa = 11.84QNKK63 pKa = 9.18KK64 pKa = 9.27FSLVVRR70 pKa = 11.84FPFKK74 pKa = 10.65RR75 pKa = 11.84SLDD78 pKa = 3.82SLNTADD84 pKa = 5.13IQMLLDD90 pKa = 4.9KK91 pKa = 10.71IADD94 pKa = 3.85PSSTGDD100 pKa = 3.48IIEE103 pKa = 3.99QLQYY107 pKa = 10.66TEE109 pKa = 4.49EE110 pKa = 4.27EE111 pKa = 4.3FVQVLEE117 pKa = 4.24EE118 pKa = 4.0QNNNKK123 pKa = 9.66DD124 pKa = 3.74LNISKK129 pKa = 8.94TINKK133 pKa = 9.3ILFDD137 pKa = 4.1VEE139 pKa = 4.99SDD141 pKa = 3.93DD142 pKa = 5.13EE143 pKa = 4.21PVEE146 pKa = 4.86CEE148 pKa = 3.5ICQEE152 pKa = 4.15TLRR155 pKa = 11.84EE156 pKa = 4.05DD157 pKa = 4.99EE158 pKa = 4.75IDD160 pKa = 4.01CGCDD164 pKa = 2.76SIEE167 pKa = 3.64IHH169 pKa = 6.51RR170 pKa = 11.84ADD172 pKa = 4.75FEE174 pKa = 4.88GFSCDD179 pKa = 5.02CPICLSDD186 pKa = 3.64KK187 pKa = 10.42HH188 pKa = 6.25AHH190 pKa = 5.71TYY192 pKa = 9.55EE193 pKa = 3.99EE194 pKa = 4.36PRR196 pKa = 11.84VEE198 pKa = 4.45EE199 pKa = 4.14EE200 pKa = 3.74PLYY203 pKa = 11.03LCEE206 pKa = 4.34CAKK209 pKa = 10.82CLGQEE214 pKa = 3.55VDD216 pKa = 3.56EE217 pKa = 4.53PLNIIEE223 pKa = 4.45EE224 pKa = 4.38TDD226 pKa = 3.99CKK228 pKa = 10.97CEE230 pKa = 4.07SCVKK234 pKa = 10.51GFDD237 pKa = 4.16DD238 pKa = 4.74IKK240 pKa = 10.81PIIIDD245 pKa = 3.6EE246 pKa = 4.56PTTQDD251 pKa = 4.29YY252 pKa = 8.65PCHH255 pKa = 6.0CQNNQQSNAQLIQTYY270 pKa = 9.55FDD272 pKa = 3.52EE273 pKa = 5.21SLYY276 pKa = 11.23SCNCVDD282 pKa = 4.61CASEE286 pKa = 4.01QKK288 pKa = 10.2QVEE291 pKa = 4.42EE292 pKa = 4.71FVDD295 pKa = 4.04EE296 pKa = 4.41PEE298 pKa = 4.35IKK300 pKa = 10.69VGTLLSNSCKK310 pKa = 10.27GVRR313 pKa = 11.84CPISKK318 pKa = 10.52EE319 pKa = 4.04EE320 pKa = 4.15TSDD323 pKa = 3.57EE324 pKa = 4.33PKK326 pKa = 10.49TEE328 pKa = 3.97VLEE331 pKa = 4.09PTIDD335 pKa = 3.66EE336 pKa = 4.49SKK338 pKa = 9.47TAYY341 pKa = 8.85EE342 pKa = 4.06LPYY345 pKa = 10.59EE346 pKa = 4.59CFCHH350 pKa = 6.67EE351 pKa = 4.55CQNPVPSNLDD361 pKa = 3.03YY362 pKa = 10.96FLWFKK367 pKa = 10.73QSQPKK372 pKa = 9.76QIDD375 pKa = 3.24YY376 pKa = 8.38TLPYY380 pKa = 9.96EE381 pKa = 4.63CQCHH385 pKa = 5.72LCSGQEE391 pKa = 3.79IEE393 pKa = 5.83LSLDD397 pKa = 3.5YY398 pKa = 10.84ILAEE402 pKa = 3.73TSVNKK407 pKa = 9.92EE408 pKa = 3.58QLEE411 pKa = 4.26EE412 pKa = 3.92VLIADD417 pKa = 4.12EE418 pKa = 5.3PIIVDD423 pKa = 3.44NFEE426 pKa = 4.4SKK428 pKa = 10.9SEE430 pKa = 3.88ILCNCDD436 pKa = 4.08HH437 pKa = 6.81NEE439 pKa = 3.92CDD441 pKa = 4.57EE442 pKa = 4.52PQLDD446 pKa = 4.22PIVNLDD452 pKa = 3.91HH453 pKa = 7.35LNAPLLAPTTTATNFEE469 pKa = 4.73CPACQHH475 pKa = 5.94VATCDD480 pKa = 3.26FCQKK484 pKa = 10.51YY485 pKa = 10.19KK486 pKa = 11.02LYY488 pKa = 10.39LLEE491 pKa = 6.12AIEE494 pKa = 5.5CPGCQKK500 pKa = 10.71LFADD504 pKa = 4.28RR505 pKa = 11.84GISIEE510 pKa = 4.32EE511 pKa = 3.92LLKK514 pKa = 10.95SYY516 pKa = 10.13FPHH519 pKa = 7.22ISYY522 pKa = 10.41DD523 pKa = 3.82GKK525 pKa = 10.71SHH527 pKa = 7.0GSQLCSLSTSEE538 pKa = 5.08QEE540 pKa = 4.72CPLAKK545 pKa = 10.6LKK547 pKa = 10.33DD548 pKa = 3.51QKK550 pKa = 11.11FIDD553 pKa = 3.63LHH555 pKa = 6.9DD556 pKa = 4.22EE557 pKa = 3.91NRR559 pKa = 11.84GIIYY563 pKa = 9.3TPEE566 pKa = 5.76DD567 pKa = 3.83GVSSGDD573 pKa = 3.47YY574 pKa = 11.11DD575 pKa = 3.68EE576 pKa = 6.01LLEE579 pKa = 4.83GNVNTKK585 pKa = 10.02RR586 pKa = 11.84GYY588 pKa = 10.36GGGGVGGPATGQGVGRR604 pKa = 11.84FAVDD608 pKa = 3.25DD609 pKa = 3.9TSAIIGAVSEE619 pKa = 4.25KK620 pKa = 10.67NKK622 pKa = 9.91PDD624 pKa = 3.5EE625 pKa = 4.78EE626 pKa = 4.39IVLVTTKK633 pKa = 9.54KK634 pKa = 8.88TGFWGIPAYY643 pKa = 9.81VLWFIVFALIAAIIIIVVMFFVFGSGLII671 pKa = 3.78
Molecular weight: 75.67 kDa
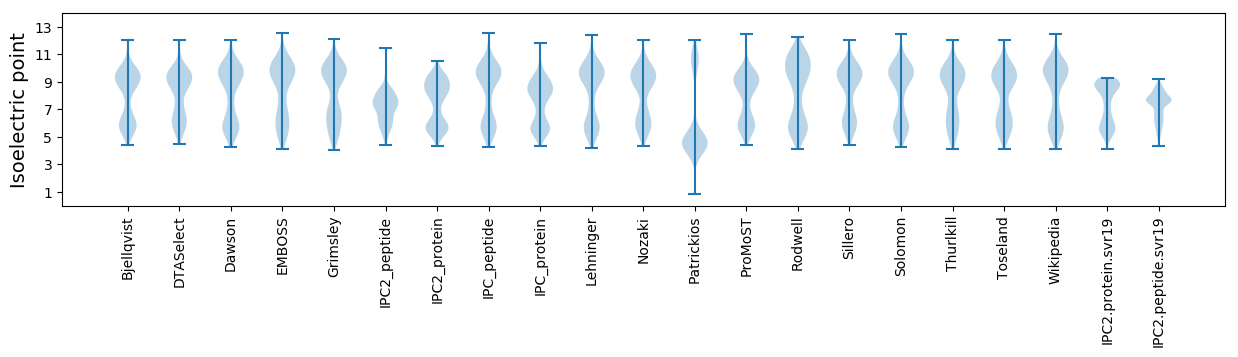
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W5V0R2|W5V0R2_9MOLU Putative phase variable surface protein VpbC OS=Mycoplasma bovoculi M165/69 OX=743966 GN=vpbC PE=4 SV=1
MM1 pKa = 7.49ALKK4 pKa = 10.12HH5 pKa = 5.7YY6 pKa = 10.68KK7 pKa = 9.88PVTNGRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.13MSALNYY21 pKa = 10.01RR22 pKa = 11.84EE23 pKa = 4.25NLTTDD28 pKa = 3.35RR29 pKa = 11.84PEE31 pKa = 4.01KK32 pKa = 10.57SLVVILKK39 pKa = 9.92KK40 pKa = 10.26HH41 pKa = 5.57SGRR44 pKa = 11.84NNQGKK49 pKa = 8.05ITTRR53 pKa = 11.84HH54 pKa = 4.31QGGRR58 pKa = 11.84QKK60 pKa = 10.67RR61 pKa = 11.84KK62 pKa = 9.61YY63 pKa = 10.33RR64 pKa = 11.84LIDD67 pKa = 3.67FKK69 pKa = 11.14RR70 pKa = 11.84NKK72 pKa = 10.2DD73 pKa = 3.69NIVAIVKK80 pKa = 9.09TIEE83 pKa = 3.71YY84 pKa = 10.37DD85 pKa = 3.49PNRR88 pKa = 11.84SANIALVSYY97 pKa = 10.98KK98 pKa = 10.57DD99 pKa = 3.44GEE101 pKa = 4.16KK102 pKa = 10.29RR103 pKa = 11.84YY104 pKa = 9.94ILAPKK109 pKa = 9.29GLQVGQQVISGDD121 pKa = 3.58NVDD124 pKa = 3.69ILVGNTLPLANIPEE138 pKa = 4.48GTFVHH143 pKa = 6.69NIEE146 pKa = 4.36LHH148 pKa = 6.19PGAGGQLMRR157 pKa = 11.84SAGASAQIQGRR168 pKa = 11.84DD169 pKa = 3.17EE170 pKa = 4.47DD171 pKa = 3.91GKK173 pKa = 11.17YY174 pKa = 10.87VILKK178 pKa = 9.66LKK180 pKa = 9.58SGEE183 pKa = 3.79YY184 pKa = 9.85RR185 pKa = 11.84RR186 pKa = 11.84ILARR190 pKa = 11.84CRR192 pKa = 11.84ATIGVVGNEE201 pKa = 3.79EE202 pKa = 3.93HH203 pKa = 6.97SLVNIGKK210 pKa = 9.86AGRR213 pKa = 11.84SRR215 pKa = 11.84HH216 pKa = 5.5LGIRR220 pKa = 11.84PTVRR224 pKa = 11.84GSVMNPNDD232 pKa = 3.69HH233 pKa = 6.5PHH235 pKa = 6.6GGGEE239 pKa = 4.24GKK241 pKa = 10.04QPIGRR246 pKa = 11.84KK247 pKa = 9.29SPLTPWGKK255 pKa = 10.11KK256 pKa = 9.7ALGVKK261 pKa = 7.53TRR263 pKa = 11.84RR264 pKa = 11.84NKK266 pKa = 10.08KK267 pKa = 10.26ASTKK271 pKa = 10.47LIIRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84KK278 pKa = 9.64DD279 pKa = 3.11SKK281 pKa = 11.34
MM1 pKa = 7.49ALKK4 pKa = 10.12HH5 pKa = 5.7YY6 pKa = 10.68KK7 pKa = 9.88PVTNGRR13 pKa = 11.84RR14 pKa = 11.84HH15 pKa = 5.13MSALNYY21 pKa = 10.01RR22 pKa = 11.84EE23 pKa = 4.25NLTTDD28 pKa = 3.35RR29 pKa = 11.84PEE31 pKa = 4.01KK32 pKa = 10.57SLVVILKK39 pKa = 9.92KK40 pKa = 10.26HH41 pKa = 5.57SGRR44 pKa = 11.84NNQGKK49 pKa = 8.05ITTRR53 pKa = 11.84HH54 pKa = 4.31QGGRR58 pKa = 11.84QKK60 pKa = 10.67RR61 pKa = 11.84KK62 pKa = 9.61YY63 pKa = 10.33RR64 pKa = 11.84LIDD67 pKa = 3.67FKK69 pKa = 11.14RR70 pKa = 11.84NKK72 pKa = 10.2DD73 pKa = 3.69NIVAIVKK80 pKa = 9.09TIEE83 pKa = 3.71YY84 pKa = 10.37DD85 pKa = 3.49PNRR88 pKa = 11.84SANIALVSYY97 pKa = 10.98KK98 pKa = 10.57DD99 pKa = 3.44GEE101 pKa = 4.16KK102 pKa = 10.29RR103 pKa = 11.84YY104 pKa = 9.94ILAPKK109 pKa = 9.29GLQVGQQVISGDD121 pKa = 3.58NVDD124 pKa = 3.69ILVGNTLPLANIPEE138 pKa = 4.48GTFVHH143 pKa = 6.69NIEE146 pKa = 4.36LHH148 pKa = 6.19PGAGGQLMRR157 pKa = 11.84SAGASAQIQGRR168 pKa = 11.84DD169 pKa = 3.17EE170 pKa = 4.47DD171 pKa = 3.91GKK173 pKa = 11.17YY174 pKa = 10.87VILKK178 pKa = 9.66LKK180 pKa = 9.58SGEE183 pKa = 3.79YY184 pKa = 9.85RR185 pKa = 11.84RR186 pKa = 11.84ILARR190 pKa = 11.84CRR192 pKa = 11.84ATIGVVGNEE201 pKa = 3.79EE202 pKa = 3.93HH203 pKa = 6.97SLVNIGKK210 pKa = 9.86AGRR213 pKa = 11.84SRR215 pKa = 11.84HH216 pKa = 5.5LGIRR220 pKa = 11.84PTVRR224 pKa = 11.84GSVMNPNDD232 pKa = 3.69HH233 pKa = 6.5PHH235 pKa = 6.6GGGEE239 pKa = 4.24GKK241 pKa = 10.04QPIGRR246 pKa = 11.84KK247 pKa = 9.29SPLTPWGKK255 pKa = 10.11KK256 pKa = 9.7ALGVKK261 pKa = 7.53TRR263 pKa = 11.84RR264 pKa = 11.84NKK266 pKa = 10.08KK267 pKa = 10.26ASTKK271 pKa = 10.47LIIRR275 pKa = 11.84RR276 pKa = 11.84RR277 pKa = 11.84KK278 pKa = 9.64DD279 pKa = 3.11SKK281 pKa = 11.34
Molecular weight: 31.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
230652 |
37 |
3779 |
398.4 |
45.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.559 ± 0.115 | 0.407 ± 0.028 |
5.424 ± 0.091 | 6.049 ± 0.114 |
6.154 ± 0.09 | 4.938 ± 0.098 |
1.411 ± 0.035 | 9.072 ± 0.127 |
9.777 ± 0.11 | 9.298 ± 0.094 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.37 ± 0.042 | 7.514 ± 0.121 |
2.992 ± 0.055 | 4.55 ± 0.067 |
2.643 ± 0.06 | 7.38 ± 0.08 |
5.367 ± 0.125 | 5.554 ± 0.074 |
0.928 ± 0.03 | 3.615 ± 0.064 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
