
Roseburia sp. CAG:18
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Roseburia; environmental samples
Average proteome isoelectric point is 6.06
Get precalculated fractions of proteins
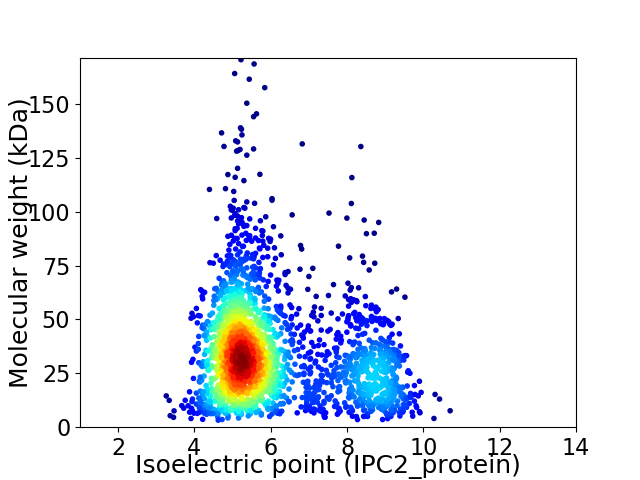
Virtual 2D-PAGE plot for 2378 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
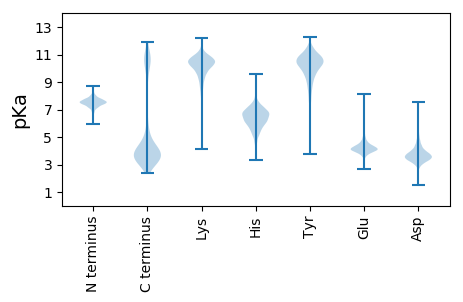
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R5UKF7|R5UKF7_9FIRM Phosphoenolpyruvate-protein phosphotransferase OS=Roseburia sp. CAG:18 OX=1262941 GN=BN518_02038 PE=3 SV=1
MM1 pKa = 7.41KK2 pKa = 10.3KK3 pKa = 10.38KK4 pKa = 10.35IISTLLCTAMLATMVAGCGVEE25 pKa = 4.19KK26 pKa = 10.82SDD28 pKa = 5.1GGSDD32 pKa = 3.79TEE34 pKa = 4.74GSASSVAEE42 pKa = 3.98MKK44 pKa = 9.97GTGDD48 pKa = 3.46NEE50 pKa = 4.12INILIYY56 pKa = 10.74AQDD59 pKa = 3.59HH60 pKa = 6.05EE61 pKa = 4.52KK62 pKa = 10.74AVYY65 pKa = 9.89QEE67 pKa = 5.33LIDD70 pKa = 4.56KK71 pKa = 7.62FTKK74 pKa = 9.74EE75 pKa = 3.93HH76 pKa = 6.75ADD78 pKa = 3.76EE79 pKa = 4.53ISTVNFEE86 pKa = 3.94VTTQDD91 pKa = 2.86EE92 pKa = 5.24YY93 pKa = 9.91ATKK96 pKa = 8.37MTAAMTAGEE105 pKa = 4.84LPDD108 pKa = 3.48IFYY111 pKa = 10.84VGPEE115 pKa = 3.86AVRR118 pKa = 11.84SYY120 pKa = 11.46VDD122 pKa = 3.17NGYY125 pKa = 9.07VQPLDD130 pKa = 4.06DD131 pKa = 5.43LVDD134 pKa = 3.65ATAVDD139 pKa = 4.26NLWPAIKK146 pKa = 9.92SAYY149 pKa = 9.37MYY151 pKa = 10.28DD152 pKa = 3.37GSNIGSGSLYY162 pKa = 10.36CLPKK166 pKa = 10.68DD167 pKa = 3.88LSCFAFAYY175 pKa = 10.69NKK177 pKa = 10.62DD178 pKa = 3.75LFDD181 pKa = 4.1EE182 pKa = 5.31AGLEE186 pKa = 4.3YY187 pKa = 9.9PDD189 pKa = 5.09PEE191 pKa = 4.6NPYY194 pKa = 8.65TWEE197 pKa = 3.93EE198 pKa = 3.97FADD201 pKa = 3.88VCQKK205 pKa = 9.03LTKK208 pKa = 10.73DD209 pKa = 3.24KK210 pKa = 11.46DD211 pKa = 3.58GDD213 pKa = 4.06GEE215 pKa = 4.05IDD217 pKa = 3.06QWGVANANAFGFTPYY232 pKa = 10.64VYY234 pKa = 10.99GNGGQFLNDD243 pKa = 3.73DD244 pKa = 3.66QTKK247 pKa = 9.82VVIDD251 pKa = 3.74EE252 pKa = 4.14NQNFKK257 pKa = 11.09DD258 pKa = 3.19AFQYY262 pKa = 9.42YY263 pKa = 9.86TDD265 pKa = 3.96LTLKK269 pKa = 10.75YY270 pKa = 9.69KK271 pKa = 8.47VTPTVEE277 pKa = 3.57QDD279 pKa = 3.24TALGGYY285 pKa = 7.23QRR287 pKa = 11.84WLDD290 pKa = 3.8GQVGFYY296 pKa = 10.98ACGTWDD302 pKa = 3.1VAAFMDD308 pKa = 5.52DD309 pKa = 3.08ATFPYY314 pKa = 10.52NWDD317 pKa = 3.37LCGWPVGPNGDD328 pKa = 3.81GKK330 pKa = 9.44STAWLGTVGFAVSSQAKK347 pKa = 9.95NPEE350 pKa = 3.86LCAEE354 pKa = 5.45LIQSLSTDD362 pKa = 3.12LDD364 pKa = 3.99GQKK367 pKa = 9.85EE368 pKa = 4.43LCGEE372 pKa = 4.18TSGKK376 pKa = 9.97SLQIPNIMDD385 pKa = 3.53YY386 pKa = 11.33AQTTFKK392 pKa = 11.28DD393 pKa = 3.45KK394 pKa = 11.56VNDD397 pKa = 3.67GTIPYY402 pKa = 9.46ASNVDD407 pKa = 3.82VIFGYY412 pKa = 10.21IEE414 pKa = 4.88GSDD417 pKa = 3.38KK418 pKa = 11.3YY419 pKa = 11.0KK420 pKa = 11.16GIFTEE425 pKa = 4.1TTYY428 pKa = 10.53TYY430 pKa = 11.18NSEE433 pKa = 3.51WWDD436 pKa = 3.55TFLEE440 pKa = 4.46GMPNVLTGEE449 pKa = 4.2VSVDD453 pKa = 3.77DD454 pKa = 3.94YY455 pKa = 11.49CKK457 pKa = 10.11QVAPKK462 pKa = 9.07MQEE465 pKa = 3.89ALDD468 pKa = 3.95NAIEE472 pKa = 4.07LQNAATNNN480 pKa = 3.58
MM1 pKa = 7.41KK2 pKa = 10.3KK3 pKa = 10.38KK4 pKa = 10.35IISTLLCTAMLATMVAGCGVEE25 pKa = 4.19KK26 pKa = 10.82SDD28 pKa = 5.1GGSDD32 pKa = 3.79TEE34 pKa = 4.74GSASSVAEE42 pKa = 3.98MKK44 pKa = 9.97GTGDD48 pKa = 3.46NEE50 pKa = 4.12INILIYY56 pKa = 10.74AQDD59 pKa = 3.59HH60 pKa = 6.05EE61 pKa = 4.52KK62 pKa = 10.74AVYY65 pKa = 9.89QEE67 pKa = 5.33LIDD70 pKa = 4.56KK71 pKa = 7.62FTKK74 pKa = 9.74EE75 pKa = 3.93HH76 pKa = 6.75ADD78 pKa = 3.76EE79 pKa = 4.53ISTVNFEE86 pKa = 3.94VTTQDD91 pKa = 2.86EE92 pKa = 5.24YY93 pKa = 9.91ATKK96 pKa = 8.37MTAAMTAGEE105 pKa = 4.84LPDD108 pKa = 3.48IFYY111 pKa = 10.84VGPEE115 pKa = 3.86AVRR118 pKa = 11.84SYY120 pKa = 11.46VDD122 pKa = 3.17NGYY125 pKa = 9.07VQPLDD130 pKa = 4.06DD131 pKa = 5.43LVDD134 pKa = 3.65ATAVDD139 pKa = 4.26NLWPAIKK146 pKa = 9.92SAYY149 pKa = 9.37MYY151 pKa = 10.28DD152 pKa = 3.37GSNIGSGSLYY162 pKa = 10.36CLPKK166 pKa = 10.68DD167 pKa = 3.88LSCFAFAYY175 pKa = 10.69NKK177 pKa = 10.62DD178 pKa = 3.75LFDD181 pKa = 4.1EE182 pKa = 5.31AGLEE186 pKa = 4.3YY187 pKa = 9.9PDD189 pKa = 5.09PEE191 pKa = 4.6NPYY194 pKa = 8.65TWEE197 pKa = 3.93EE198 pKa = 3.97FADD201 pKa = 3.88VCQKK205 pKa = 9.03LTKK208 pKa = 10.73DD209 pKa = 3.24KK210 pKa = 11.46DD211 pKa = 3.58GDD213 pKa = 4.06GEE215 pKa = 4.05IDD217 pKa = 3.06QWGVANANAFGFTPYY232 pKa = 10.64VYY234 pKa = 10.99GNGGQFLNDD243 pKa = 3.73DD244 pKa = 3.66QTKK247 pKa = 9.82VVIDD251 pKa = 3.74EE252 pKa = 4.14NQNFKK257 pKa = 11.09DD258 pKa = 3.19AFQYY262 pKa = 9.42YY263 pKa = 9.86TDD265 pKa = 3.96LTLKK269 pKa = 10.75YY270 pKa = 9.69KK271 pKa = 8.47VTPTVEE277 pKa = 3.57QDD279 pKa = 3.24TALGGYY285 pKa = 7.23QRR287 pKa = 11.84WLDD290 pKa = 3.8GQVGFYY296 pKa = 10.98ACGTWDD302 pKa = 3.1VAAFMDD308 pKa = 5.52DD309 pKa = 3.08ATFPYY314 pKa = 10.52NWDD317 pKa = 3.37LCGWPVGPNGDD328 pKa = 3.81GKK330 pKa = 9.44STAWLGTVGFAVSSQAKK347 pKa = 9.95NPEE350 pKa = 3.86LCAEE354 pKa = 5.45LIQSLSTDD362 pKa = 3.12LDD364 pKa = 3.99GQKK367 pKa = 9.85EE368 pKa = 4.43LCGEE372 pKa = 4.18TSGKK376 pKa = 9.97SLQIPNIMDD385 pKa = 3.53YY386 pKa = 11.33AQTTFKK392 pKa = 11.28DD393 pKa = 3.45KK394 pKa = 11.56VNDD397 pKa = 3.67GTIPYY402 pKa = 9.46ASNVDD407 pKa = 3.82VIFGYY412 pKa = 10.21IEE414 pKa = 4.88GSDD417 pKa = 3.38KK418 pKa = 11.3YY419 pKa = 11.0KK420 pKa = 11.16GIFTEE425 pKa = 4.1TTYY428 pKa = 10.53TYY430 pKa = 11.18NSEE433 pKa = 3.51WWDD436 pKa = 3.55TFLEE440 pKa = 4.46GMPNVLTGEE449 pKa = 4.2VSVDD453 pKa = 3.77DD454 pKa = 3.94YY455 pKa = 11.49CKK457 pKa = 10.11QVAPKK462 pKa = 9.07MQEE465 pKa = 3.89ALDD468 pKa = 3.95NAIEE472 pKa = 4.07LQNAATNNN480 pKa = 3.58
Molecular weight: 52.85 kDa
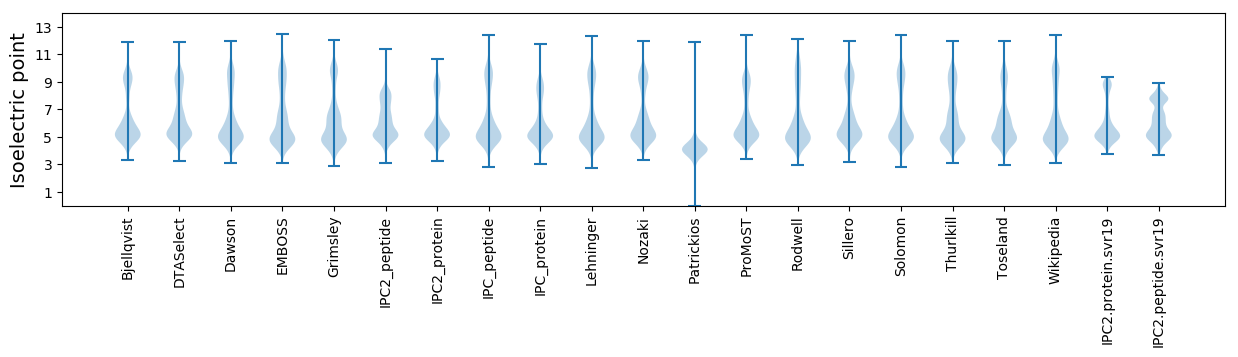
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R5UKR8|R5UKR8_9FIRM ABC transmembrane type-1 domain-containing protein OS=Roseburia sp. CAG:18 OX=1262941 GN=BN518_02146 PE=3 SV=1
MM1 pKa = 7.51HH2 pKa = 7.22KK3 pKa = 10.29HH4 pKa = 5.1FGKK7 pKa = 8.85RR8 pKa = 11.84TKK10 pKa = 9.99KK11 pKa = 9.18KK12 pKa = 8.44HH13 pKa = 6.29HH14 pKa = 6.67IVLICSAVVIAAFVLRR30 pKa = 11.84MPVEE34 pKa = 4.11MNVADD39 pKa = 3.98ACGMQQKK46 pKa = 10.35VNVLSSSMHH55 pKa = 5.13NRR57 pKa = 11.84QAEE60 pKa = 4.33DD61 pKa = 3.62YY62 pKa = 10.53LNDD65 pKa = 3.78AGEE68 pKa = 4.25FCISQTVLRR77 pKa = 11.84EE78 pKa = 4.22GKK80 pKa = 9.59PSVVPQVRR88 pKa = 11.84RR89 pKa = 11.84IGLRR93 pKa = 11.84FLPVSHH99 pKa = 6.34MTQQWLLVQRR109 pKa = 11.84ILTSKK114 pKa = 10.41PKK116 pKa = 10.48FLVPPARR123 pKa = 11.84RR124 pKa = 11.84LIVGRR129 pKa = 11.84YY130 pKa = 7.6IRR132 pKa = 11.84ANPVCVV138 pKa = 3.24
MM1 pKa = 7.51HH2 pKa = 7.22KK3 pKa = 10.29HH4 pKa = 5.1FGKK7 pKa = 8.85RR8 pKa = 11.84TKK10 pKa = 9.99KK11 pKa = 9.18KK12 pKa = 8.44HH13 pKa = 6.29HH14 pKa = 6.67IVLICSAVVIAAFVLRR30 pKa = 11.84MPVEE34 pKa = 4.11MNVADD39 pKa = 3.98ACGMQQKK46 pKa = 10.35VNVLSSSMHH55 pKa = 5.13NRR57 pKa = 11.84QAEE60 pKa = 4.33DD61 pKa = 3.62YY62 pKa = 10.53LNDD65 pKa = 3.78AGEE68 pKa = 4.25FCISQTVLRR77 pKa = 11.84EE78 pKa = 4.22GKK80 pKa = 9.59PSVVPQVRR88 pKa = 11.84RR89 pKa = 11.84IGLRR93 pKa = 11.84FLPVSHH99 pKa = 6.34MTQQWLLVQRR109 pKa = 11.84ILTSKK114 pKa = 10.41PKK116 pKa = 10.48FLVPPARR123 pKa = 11.84RR124 pKa = 11.84LIVGRR129 pKa = 11.84YY130 pKa = 7.6IRR132 pKa = 11.84ANPVCVV138 pKa = 3.24
Molecular weight: 15.68 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
763030 |
30 |
1517 |
320.9 |
35.99 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.654 ± 0.05 | 1.491 ± 0.019 |
5.822 ± 0.046 | 7.312 ± 0.053 |
3.984 ± 0.035 | 6.995 ± 0.045 |
1.926 ± 0.022 | 7.119 ± 0.048 |
6.954 ± 0.047 | 8.693 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.238 ± 0.028 | 4.31 ± 0.04 |
3.201 ± 0.03 | 3.62 ± 0.034 |
4.286 ± 0.034 | 5.627 ± 0.041 |
5.61 ± 0.037 | 7.092 ± 0.042 |
0.893 ± 0.019 | 4.171 ± 0.035 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
