
Wenzhou tombus-like virus 7
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.63
Get precalculated fractions of proteins
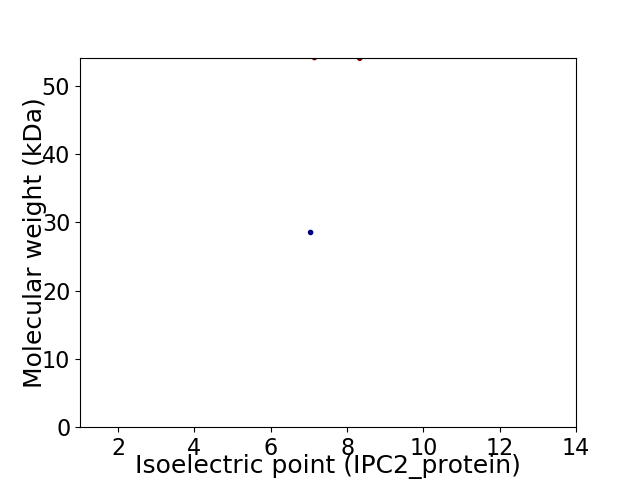
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KH28|A0A1L3KH28_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 7 OX=1923677 PE=4 SV=1
MM1 pKa = 7.76RR2 pKa = 11.84SVLLGTDD9 pKa = 3.04SDD11 pKa = 4.0NVVITKK17 pKa = 7.62EE18 pKa = 4.05FPVFYY23 pKa = 10.12SGYY26 pKa = 9.65PFTDD30 pKa = 3.99DD31 pKa = 3.93NVSTQTDD38 pKa = 3.84VVPIEE43 pKa = 4.11VDD45 pKa = 3.33LARR48 pKa = 11.84AMNEE52 pKa = 4.04SLFSYY57 pKa = 10.57SPGISIDD64 pKa = 3.4NRR66 pKa = 11.84HH67 pKa = 5.48TPRR70 pKa = 11.84YY71 pKa = 9.01DD72 pKa = 3.81QIDD75 pKa = 3.5ATLWQWLCPPRR86 pKa = 11.84LRR88 pKa = 11.84GARR91 pKa = 11.84LLLSNRR97 pKa = 11.84EE98 pKa = 3.86TYY100 pKa = 10.32AYY102 pKa = 9.79EE103 pKa = 4.0LLKK106 pKa = 11.0ALNDD110 pKa = 5.02PISEE114 pKa = 4.6TIDD117 pKa = 3.13EE118 pKa = 4.55ALYY121 pKa = 10.85KK122 pKa = 10.44HH123 pKa = 6.43GNLIKK128 pKa = 9.81TAVDD132 pKa = 3.17YY133 pKa = 10.99LYY135 pKa = 10.41TFRR138 pKa = 11.84ATMPGVQFTHH148 pKa = 7.34DD149 pKa = 3.91KK150 pKa = 11.53DD151 pKa = 3.87EE152 pKa = 4.16MGMLIVRR159 pKa = 11.84ADD161 pKa = 3.01AWISIKK167 pKa = 10.65LKK169 pKa = 9.81EE170 pKa = 4.17VLIEE174 pKa = 4.03RR175 pKa = 11.84PNFRR179 pKa = 11.84GMHH182 pKa = 4.49MQFIKK187 pKa = 10.38MIFLLLCQQKK197 pKa = 10.55SFLDD201 pKa = 3.73EE202 pKa = 4.09MSSDD206 pKa = 4.11LYY208 pKa = 11.03HH209 pKa = 7.4SRR211 pKa = 11.84TLSLNARR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84NGYY224 pKa = 9.59RR225 pKa = 11.84KK226 pKa = 9.93YY227 pKa = 10.32PWLGWFSPLANRR239 pKa = 11.84PEE241 pKa = 4.14PSRR244 pKa = 11.84AA245 pKa = 3.29
MM1 pKa = 7.76RR2 pKa = 11.84SVLLGTDD9 pKa = 3.04SDD11 pKa = 4.0NVVITKK17 pKa = 7.62EE18 pKa = 4.05FPVFYY23 pKa = 10.12SGYY26 pKa = 9.65PFTDD30 pKa = 3.99DD31 pKa = 3.93NVSTQTDD38 pKa = 3.84VVPIEE43 pKa = 4.11VDD45 pKa = 3.33LARR48 pKa = 11.84AMNEE52 pKa = 4.04SLFSYY57 pKa = 10.57SPGISIDD64 pKa = 3.4NRR66 pKa = 11.84HH67 pKa = 5.48TPRR70 pKa = 11.84YY71 pKa = 9.01DD72 pKa = 3.81QIDD75 pKa = 3.5ATLWQWLCPPRR86 pKa = 11.84LRR88 pKa = 11.84GARR91 pKa = 11.84LLLSNRR97 pKa = 11.84EE98 pKa = 3.86TYY100 pKa = 10.32AYY102 pKa = 9.79EE103 pKa = 4.0LLKK106 pKa = 11.0ALNDD110 pKa = 5.02PISEE114 pKa = 4.6TIDD117 pKa = 3.13EE118 pKa = 4.55ALYY121 pKa = 10.85KK122 pKa = 10.44HH123 pKa = 6.43GNLIKK128 pKa = 9.81TAVDD132 pKa = 3.17YY133 pKa = 10.99LYY135 pKa = 10.41TFRR138 pKa = 11.84ATMPGVQFTHH148 pKa = 7.34DD149 pKa = 3.91KK150 pKa = 11.53DD151 pKa = 3.87EE152 pKa = 4.16MGMLIVRR159 pKa = 11.84ADD161 pKa = 3.01AWISIKK167 pKa = 10.65LKK169 pKa = 9.81EE170 pKa = 4.17VLIEE174 pKa = 4.03RR175 pKa = 11.84PNFRR179 pKa = 11.84GMHH182 pKa = 4.49MQFIKK187 pKa = 10.38MIFLLLCQQKK197 pKa = 10.55SFLDD201 pKa = 3.73EE202 pKa = 4.09MSSDD206 pKa = 4.11LYY208 pKa = 11.03HH209 pKa = 7.4SRR211 pKa = 11.84TLSLNARR218 pKa = 11.84RR219 pKa = 11.84RR220 pKa = 11.84RR221 pKa = 11.84NGYY224 pKa = 9.59RR225 pKa = 11.84KK226 pKa = 9.93YY227 pKa = 10.32PWLGWFSPLANRR239 pKa = 11.84PEE241 pKa = 4.14PSRR244 pKa = 11.84AA245 pKa = 3.29
Molecular weight: 28.51 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KH28|A0A1L3KH28_9VIRU RNA-directed RNA polymerase OS=Wenzhou tombus-like virus 7 OX=1923677 PE=4 SV=1
MM1 pKa = 7.74TYY3 pKa = 10.38QPGIDD8 pKa = 3.47VLVRR12 pKa = 11.84AISEE16 pKa = 3.86RR17 pKa = 11.84LYY19 pKa = 11.03CINTPNGWIQPLVPKK34 pKa = 10.63RR35 pKa = 11.84EE36 pKa = 4.25DD37 pKa = 3.37VSGLALLFDD46 pKa = 5.0AARR49 pKa = 11.84QHH51 pKa = 6.56IRR53 pKa = 11.84VQHH56 pKa = 5.73PMSAEE61 pKa = 3.83QFANCYY67 pKa = 7.26TGPRR71 pKa = 11.84KK72 pKa = 9.46RR73 pKa = 11.84RR74 pKa = 11.84YY75 pKa = 8.74LAAARR80 pKa = 11.84SLAEE84 pKa = 4.37SGISHH89 pKa = 6.34QDD91 pKa = 2.74ARR93 pKa = 11.84LKK95 pKa = 11.03YY96 pKa = 8.17FLKK99 pKa = 10.79FEE101 pKa = 4.45TYY103 pKa = 10.93NFDD106 pKa = 3.79EE107 pKa = 5.16KK108 pKa = 10.94PNPSPRR114 pKa = 11.84GINPRR119 pKa = 11.84SDD121 pKa = 3.23RR122 pKa = 11.84YY123 pKa = 10.28LVEE126 pKa = 4.48LGRR129 pKa = 11.84FLHH132 pKa = 6.6SIEE135 pKa = 3.71KK136 pKa = 10.14RR137 pKa = 11.84IYY139 pKa = 9.5KK140 pKa = 10.37AIALVFGHH148 pKa = 5.91PVIMKK153 pKa = 9.79GYY155 pKa = 8.59NQAQRR160 pKa = 11.84GAILDD165 pKa = 3.79KK166 pKa = 11.23LMSEE170 pKa = 4.15VDD172 pKa = 3.99DD173 pKa = 4.56PVAIPADD180 pKa = 3.37ASRR183 pKa = 11.84FEE185 pKa = 3.85QSIDD189 pKa = 3.38IPHH192 pKa = 7.03LEE194 pKa = 4.6VEE196 pKa = 4.64HH197 pKa = 6.19KK198 pKa = 10.0WYY200 pKa = 11.19LEE202 pKa = 3.7FFRR205 pKa = 11.84GEE207 pKa = 4.1DD208 pKa = 3.3RR209 pKa = 11.84KK210 pKa = 10.53HH211 pKa = 6.04LAFLLDD217 pKa = 3.79LQKK220 pKa = 10.9HH221 pKa = 5.41NKK223 pKa = 9.16GSAHH227 pKa = 6.87ASDD230 pKa = 4.52GDD232 pKa = 4.04LKK234 pKa = 11.16FSIDD238 pKa = 3.96GKK240 pKa = 10.98RR241 pKa = 11.84MSGDD245 pKa = 3.34KK246 pKa = 9.43NTSSGNCIISASLWYY261 pKa = 10.06EE262 pKa = 3.7FAKK265 pKa = 10.6EE266 pKa = 4.12KK267 pKa = 10.65GWLTLTRR274 pKa = 11.84FFCDD278 pKa = 2.83GDD280 pKa = 4.06DD281 pKa = 3.38AVMIIPRR288 pKa = 11.84KK289 pKa = 8.01YY290 pKa = 10.55LKK292 pKa = 10.36EE293 pKa = 3.87FQQDD297 pKa = 3.78YY298 pKa = 11.32LVFCSKK304 pKa = 10.58RR305 pKa = 11.84GFRR308 pKa = 11.84MKK310 pKa = 10.28LDD312 pKa = 3.29KK313 pKa = 10.69VACILEE319 pKa = 4.32QIDD322 pKa = 4.0FCQSRR327 pKa = 11.84PVWTPQGYY335 pKa = 9.97VMVRR339 pKa = 11.84NPVSVSKK346 pKa = 10.47DD347 pKa = 3.53CHH349 pKa = 6.41SKK351 pKa = 11.29APINNEE357 pKa = 3.49KK358 pKa = 10.03IARR361 pKa = 11.84RR362 pKa = 11.84WIKK365 pKa = 10.68AVGLCGLSCNGGIPVMQEE383 pKa = 3.72FYY385 pKa = 10.97RR386 pKa = 11.84AMIRR390 pKa = 11.84YY391 pKa = 9.27AGDD394 pKa = 3.23VKK396 pKa = 11.11ALDD399 pKa = 3.77LTHH402 pKa = 7.19PLMVRR407 pKa = 11.84QFRR410 pKa = 11.84YY411 pKa = 9.66KK412 pKa = 10.01VYY414 pKa = 10.59GMDD417 pKa = 3.4RR418 pKa = 11.84EE419 pKa = 4.05EE420 pKa = 5.04DD421 pKa = 3.72VIHH424 pKa = 6.5PRR426 pKa = 11.84TRR428 pKa = 11.84ASYY431 pKa = 9.82AVAFNVSPSEE441 pKa = 3.76QLLLEE446 pKa = 3.91KK447 pKa = 10.76HH448 pKa = 5.36MAEE451 pKa = 3.9SHH453 pKa = 7.08LEE455 pKa = 3.95FTFEE459 pKa = 4.07TRR461 pKa = 11.84PSITRR466 pKa = 11.84IGLMM470 pKa = 3.96
MM1 pKa = 7.74TYY3 pKa = 10.38QPGIDD8 pKa = 3.47VLVRR12 pKa = 11.84AISEE16 pKa = 3.86RR17 pKa = 11.84LYY19 pKa = 11.03CINTPNGWIQPLVPKK34 pKa = 10.63RR35 pKa = 11.84EE36 pKa = 4.25DD37 pKa = 3.37VSGLALLFDD46 pKa = 5.0AARR49 pKa = 11.84QHH51 pKa = 6.56IRR53 pKa = 11.84VQHH56 pKa = 5.73PMSAEE61 pKa = 3.83QFANCYY67 pKa = 7.26TGPRR71 pKa = 11.84KK72 pKa = 9.46RR73 pKa = 11.84RR74 pKa = 11.84YY75 pKa = 8.74LAAARR80 pKa = 11.84SLAEE84 pKa = 4.37SGISHH89 pKa = 6.34QDD91 pKa = 2.74ARR93 pKa = 11.84LKK95 pKa = 11.03YY96 pKa = 8.17FLKK99 pKa = 10.79FEE101 pKa = 4.45TYY103 pKa = 10.93NFDD106 pKa = 3.79EE107 pKa = 5.16KK108 pKa = 10.94PNPSPRR114 pKa = 11.84GINPRR119 pKa = 11.84SDD121 pKa = 3.23RR122 pKa = 11.84YY123 pKa = 10.28LVEE126 pKa = 4.48LGRR129 pKa = 11.84FLHH132 pKa = 6.6SIEE135 pKa = 3.71KK136 pKa = 10.14RR137 pKa = 11.84IYY139 pKa = 9.5KK140 pKa = 10.37AIALVFGHH148 pKa = 5.91PVIMKK153 pKa = 9.79GYY155 pKa = 8.59NQAQRR160 pKa = 11.84GAILDD165 pKa = 3.79KK166 pKa = 11.23LMSEE170 pKa = 4.15VDD172 pKa = 3.99DD173 pKa = 4.56PVAIPADD180 pKa = 3.37ASRR183 pKa = 11.84FEE185 pKa = 3.85QSIDD189 pKa = 3.38IPHH192 pKa = 7.03LEE194 pKa = 4.6VEE196 pKa = 4.64HH197 pKa = 6.19KK198 pKa = 10.0WYY200 pKa = 11.19LEE202 pKa = 3.7FFRR205 pKa = 11.84GEE207 pKa = 4.1DD208 pKa = 3.3RR209 pKa = 11.84KK210 pKa = 10.53HH211 pKa = 6.04LAFLLDD217 pKa = 3.79LQKK220 pKa = 10.9HH221 pKa = 5.41NKK223 pKa = 9.16GSAHH227 pKa = 6.87ASDD230 pKa = 4.52GDD232 pKa = 4.04LKK234 pKa = 11.16FSIDD238 pKa = 3.96GKK240 pKa = 10.98RR241 pKa = 11.84MSGDD245 pKa = 3.34KK246 pKa = 9.43NTSSGNCIISASLWYY261 pKa = 10.06EE262 pKa = 3.7FAKK265 pKa = 10.6EE266 pKa = 4.12KK267 pKa = 10.65GWLTLTRR274 pKa = 11.84FFCDD278 pKa = 2.83GDD280 pKa = 4.06DD281 pKa = 3.38AVMIIPRR288 pKa = 11.84KK289 pKa = 8.01YY290 pKa = 10.55LKK292 pKa = 10.36EE293 pKa = 3.87FQQDD297 pKa = 3.78YY298 pKa = 11.32LVFCSKK304 pKa = 10.58RR305 pKa = 11.84GFRR308 pKa = 11.84MKK310 pKa = 10.28LDD312 pKa = 3.29KK313 pKa = 10.69VACILEE319 pKa = 4.32QIDD322 pKa = 4.0FCQSRR327 pKa = 11.84PVWTPQGYY335 pKa = 9.97VMVRR339 pKa = 11.84NPVSVSKK346 pKa = 10.47DD347 pKa = 3.53CHH349 pKa = 6.41SKK351 pKa = 11.29APINNEE357 pKa = 3.49KK358 pKa = 10.03IARR361 pKa = 11.84RR362 pKa = 11.84WIKK365 pKa = 10.68AVGLCGLSCNGGIPVMQEE383 pKa = 3.72FYY385 pKa = 10.97RR386 pKa = 11.84AMIRR390 pKa = 11.84YY391 pKa = 9.27AGDD394 pKa = 3.23VKK396 pKa = 11.11ALDD399 pKa = 3.77LTHH402 pKa = 7.19PLMVRR407 pKa = 11.84QFRR410 pKa = 11.84YY411 pKa = 9.66KK412 pKa = 10.01VYY414 pKa = 10.59GMDD417 pKa = 3.4RR418 pKa = 11.84EE419 pKa = 4.05EE420 pKa = 5.04DD421 pKa = 3.72VIHH424 pKa = 6.5PRR426 pKa = 11.84TRR428 pKa = 11.84ASYY431 pKa = 9.82AVAFNVSPSEE441 pKa = 3.76QLLLEE446 pKa = 3.91KK447 pKa = 10.76HH448 pKa = 5.36MAEE451 pKa = 3.9SHH453 pKa = 7.08LEE455 pKa = 3.95FTFEE459 pKa = 4.07TRR461 pKa = 11.84PSITRR466 pKa = 11.84IGLMM470 pKa = 3.96
Molecular weight: 54.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
715 |
245 |
470 |
357.5 |
41.26 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.853 ± 0.63 | 1.678 ± 0.477 |
6.294 ± 0.357 | 5.455 ± 0.308 |
4.895 ± 0.224 | 5.175 ± 0.604 |
2.797 ± 0.418 | 6.154 ± 0.243 |
5.594 ± 0.836 | 9.51 ± 1.06 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.217 ± 0.252 | 3.636 ± 0.472 |
5.315 ± 0.221 | 3.497 ± 0.353 |
7.972 ± 0.106 | 6.853 ± 0.273 |
3.776 ± 1.071 | 5.455 ± 0.308 |
1.538 ± 0.278 | 4.336 ± 0.311 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
