
Coprococcus catus
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Lachnospiraceae; Coprococcus
Average proteome isoelectric point is 6.03
Get precalculated fractions of proteins
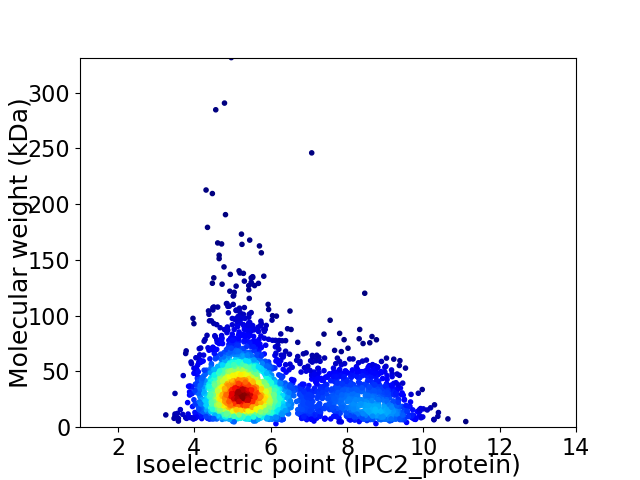
Virtual 2D-PAGE plot for 3079 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3E2XS14|A0A3E2XS14_9FIRM Stage 0 sporulation protein A homolog OS=Coprococcus catus OX=116085 GN=DW747_01320 PE=4 SV=1
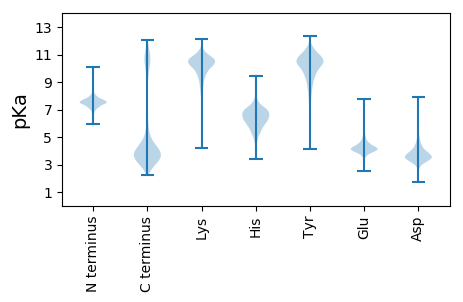
MM1 pKa = 7.68KK2 pKa = 10.31KK3 pKa = 10.06RR4 pKa = 11.84VITGLMAASVIVSLAGCGAGQKK26 pKa = 10.34EE27 pKa = 4.91SVEE30 pKa = 4.28STAPATTTEE39 pKa = 4.34TVAASQVEE47 pKa = 4.4TAEE50 pKa = 3.94ATEE53 pKa = 4.33TEE55 pKa = 4.39KK56 pKa = 10.37TVQEE60 pKa = 4.03DD61 pKa = 3.14RR62 pKa = 11.84AARR65 pKa = 11.84IKK67 pKa = 10.49AYY69 pKa = 10.0QAALDD74 pKa = 4.4NIYY77 pKa = 10.48QNHH80 pKa = 6.76ILPDD84 pKa = 3.99GSEE87 pKa = 3.99LRR89 pKa = 11.84DD90 pKa = 4.54DD91 pKa = 4.26ISGEE95 pKa = 3.69MSGNKK100 pKa = 9.14FAVYY104 pKa = 10.25DD105 pKa = 3.59VDD107 pKa = 4.21HH108 pKa = 7.9DD109 pKa = 4.04DD110 pKa = 3.99RR111 pKa = 11.84EE112 pKa = 4.32EE113 pKa = 4.52LVFCYY118 pKa = 8.97TATSMASMIAKK129 pKa = 9.89IYY131 pKa = 10.66DD132 pKa = 3.34YY133 pKa = 11.45DD134 pKa = 3.79EE135 pKa = 5.09ASGEE139 pKa = 4.12LYY141 pKa = 10.7EE142 pKa = 4.86EE143 pKa = 5.11LSQFPMLTFYY153 pKa = 11.57DD154 pKa = 3.78NATIGAGISHH164 pKa = 6.0NQGYY168 pKa = 10.48AGEE171 pKa = 4.42FWPYY175 pKa = 10.18TFYY178 pKa = 10.67TYY180 pKa = 11.16DD181 pKa = 3.55AANDD185 pKa = 3.8SYY187 pKa = 10.8RR188 pKa = 11.84AEE190 pKa = 4.28YY191 pKa = 10.07FVDD194 pKa = 4.03AWDD197 pKa = 4.08KK198 pKa = 11.27SLSEE202 pKa = 3.96VNYY205 pKa = 10.55NGEE208 pKa = 4.44AFPDD212 pKa = 4.14DD213 pKa = 4.23ADD215 pKa = 4.05KK216 pKa = 11.58DD217 pKa = 3.89GDD219 pKa = 4.17GIVYY223 pKa = 10.27YY224 pKa = 10.9LEE226 pKa = 5.49APDD229 pKa = 4.34GSEE232 pKa = 3.79TGEE235 pKa = 4.51PLDD238 pKa = 5.51KK239 pKa = 11.04DD240 pKa = 5.69AFDD243 pKa = 3.34QWISEE248 pKa = 4.23NVGKK252 pKa = 10.6EE253 pKa = 3.87EE254 pKa = 4.85NIPFQDD260 pKa = 3.53LTEE263 pKa = 4.51DD264 pKa = 4.78AISQIQQ270 pKa = 3.04
MM1 pKa = 7.68KK2 pKa = 10.31KK3 pKa = 10.06RR4 pKa = 11.84VITGLMAASVIVSLAGCGAGQKK26 pKa = 10.34EE27 pKa = 4.91SVEE30 pKa = 4.28STAPATTTEE39 pKa = 4.34TVAASQVEE47 pKa = 4.4TAEE50 pKa = 3.94ATEE53 pKa = 4.33TEE55 pKa = 4.39KK56 pKa = 10.37TVQEE60 pKa = 4.03DD61 pKa = 3.14RR62 pKa = 11.84AARR65 pKa = 11.84IKK67 pKa = 10.49AYY69 pKa = 10.0QAALDD74 pKa = 4.4NIYY77 pKa = 10.48QNHH80 pKa = 6.76ILPDD84 pKa = 3.99GSEE87 pKa = 3.99LRR89 pKa = 11.84DD90 pKa = 4.54DD91 pKa = 4.26ISGEE95 pKa = 3.69MSGNKK100 pKa = 9.14FAVYY104 pKa = 10.25DD105 pKa = 3.59VDD107 pKa = 4.21HH108 pKa = 7.9DD109 pKa = 4.04DD110 pKa = 3.99RR111 pKa = 11.84EE112 pKa = 4.32EE113 pKa = 4.52LVFCYY118 pKa = 8.97TATSMASMIAKK129 pKa = 9.89IYY131 pKa = 10.66DD132 pKa = 3.34YY133 pKa = 11.45DD134 pKa = 3.79EE135 pKa = 5.09ASGEE139 pKa = 4.12LYY141 pKa = 10.7EE142 pKa = 4.86EE143 pKa = 5.11LSQFPMLTFYY153 pKa = 11.57DD154 pKa = 3.78NATIGAGISHH164 pKa = 6.0NQGYY168 pKa = 10.48AGEE171 pKa = 4.42FWPYY175 pKa = 10.18TFYY178 pKa = 10.67TYY180 pKa = 11.16DD181 pKa = 3.55AANDD185 pKa = 3.8SYY187 pKa = 10.8RR188 pKa = 11.84AEE190 pKa = 4.28YY191 pKa = 10.07FVDD194 pKa = 4.03AWDD197 pKa = 4.08KK198 pKa = 11.27SLSEE202 pKa = 3.96VNYY205 pKa = 10.55NGEE208 pKa = 4.44AFPDD212 pKa = 4.14DD213 pKa = 4.23ADD215 pKa = 4.05KK216 pKa = 11.58DD217 pKa = 3.89GDD219 pKa = 4.17GIVYY223 pKa = 10.27YY224 pKa = 10.9LEE226 pKa = 5.49APDD229 pKa = 4.34GSEE232 pKa = 3.79TGEE235 pKa = 4.51PLDD238 pKa = 5.51KK239 pKa = 11.04DD240 pKa = 5.69AFDD243 pKa = 3.34QWISEE248 pKa = 4.23NVGKK252 pKa = 10.6EE253 pKa = 3.87EE254 pKa = 4.85NIPFQDD260 pKa = 3.53LTEE263 pKa = 4.51DD264 pKa = 4.78AISQIQQ270 pKa = 3.04
Molecular weight: 29.83 kDa
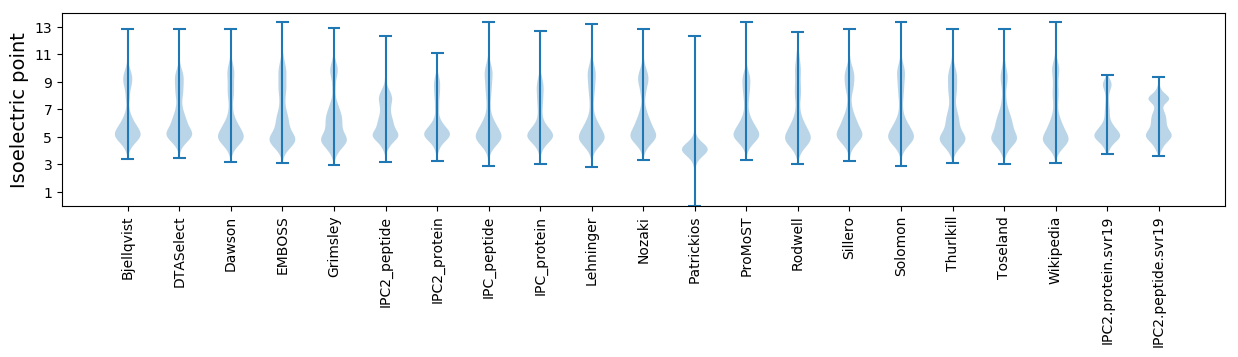
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3E2TNR7|A0A3E2TNR7_9FIRM Stp1/IreP family PP2C-type Ser/Thr phosphatase OS=Coprococcus catus OX=116085 GN=DW070_07630 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.78TRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.94VHH16 pKa = 5.99GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
MM1 pKa = 7.67KK2 pKa = 8.71MTFQPKK8 pKa = 7.78TRR10 pKa = 11.84QRR12 pKa = 11.84SKK14 pKa = 8.94VHH16 pKa = 5.99GFRR19 pKa = 11.84KK20 pKa = 10.0RR21 pKa = 11.84MSTANGRR28 pKa = 11.84KK29 pKa = 8.89VLAARR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.71GRR39 pKa = 11.84KK40 pKa = 8.73VLSAA44 pKa = 4.05
Molecular weight: 5.07 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
982488 |
27 |
2929 |
319.1 |
35.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.872 ± 0.047 | 1.522 ± 0.017 |
5.923 ± 0.035 | 7.269 ± 0.054 |
3.892 ± 0.032 | 7.022 ± 0.039 |
1.955 ± 0.023 | 7.38 ± 0.046 |
6.52 ± 0.042 | 8.837 ± 0.046 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.349 ± 0.027 | 4.245 ± 0.032 |
3.272 ± 0.024 | 3.445 ± 0.027 |
4.441 ± 0.038 | 5.734 ± 0.036 |
5.426 ± 0.038 | 6.907 ± 0.04 |
0.893 ± 0.015 | 4.095 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
