
Avian metaavulavirus 7
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Avulavirinae; Metaavulavirus
Average proteome isoelectric point is 5.99
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|C6FGZ0|C6FGZ0_9MONO Hemagglutinin-neuraminidase OS=Avian metaavulavirus 7 OX=2560317 GN=HN PE=3 SV=1
MM1 pKa = 7.92RR2 pKa = 11.84VRR4 pKa = 11.84PLIIILVLLVLLWLNILPVIGLDD27 pKa = 3.34NSKK30 pKa = 9.92IAQAGIISAQEE41 pKa = 3.78YY42 pKa = 9.53AVNVYY47 pKa = 9.97SQSNEE52 pKa = 3.37AYY54 pKa = 8.7IALRR58 pKa = 11.84TVPYY62 pKa = 10.09IPPHH66 pKa = 6.01NLSCFQDD73 pKa = 4.53LINTYY78 pKa = 8.44NTTIQNIFSPIQDD91 pKa = 3.85QITSITSASTLPSSRR106 pKa = 11.84FAGLVVGAIALGVATSAQITAAVALTKK133 pKa = 10.39AQQNAQEE140 pKa = 4.53IIRR143 pKa = 11.84LRR145 pKa = 11.84DD146 pKa = 3.66SIQNTINAVNDD157 pKa = 3.24ITVGLSSIGVALSKK171 pKa = 10.05VQNYY175 pKa = 10.03LNDD178 pKa = 3.87VINPALQNLSCQVSALNLGIQLNLYY203 pKa = 8.18LTEE206 pKa = 3.8ITTIFGPQITNPSLTPLSIQALYY229 pKa = 9.57TLAGDD234 pKa = 4.04NLMQFLTRR242 pKa = 11.84YY243 pKa = 9.77GYY245 pKa = 11.41GEE247 pKa = 4.2TSVSSILEE255 pKa = 3.98SGLISAQIVSFDD267 pKa = 3.67KK268 pKa = 10.22QTGIAILYY276 pKa = 6.9VTLPSIATLSGSRR289 pKa = 11.84VTKK292 pKa = 10.54LMSVSVQTGVGEE304 pKa = 4.46GSAIVPSYY312 pKa = 10.7VIQQGTVIEE321 pKa = 4.24EE322 pKa = 4.64FIPDD326 pKa = 3.27SCIFTRR332 pKa = 11.84SDD334 pKa = 3.71VYY336 pKa = 10.36CTQLYY341 pKa = 10.78SKK343 pKa = 10.23LLPDD347 pKa = 5.99SILQCLQGSMADD359 pKa = 3.72CQFTRR364 pKa = 11.84SLGSFANRR372 pKa = 11.84FMTVAGGVIANCQTVLCRR390 pKa = 11.84CYY392 pKa = 10.24NPVMIIPQNNGIAVTLIDD410 pKa = 4.55GSLCKK415 pKa = 10.08EE416 pKa = 4.31LEE418 pKa = 4.17LEE420 pKa = 4.73GIRR423 pKa = 11.84LTMADD428 pKa = 4.18PVFASYY434 pKa = 11.11SRR436 pKa = 11.84DD437 pKa = 3.32LIINGNQFAPSDD449 pKa = 3.72ALDD452 pKa = 3.42ISSEE456 pKa = 4.02LGQLNNSISSATDD469 pKa = 3.24NLQKK473 pKa = 10.82AQEE476 pKa = 4.31SLNKK480 pKa = 10.31SIIPAATSSWLIILLFVLVSISLVIGCISIYY511 pKa = 10.44FIYY514 pKa = 10.23KK515 pKa = 9.93HH516 pKa = 5.42STTNRR521 pKa = 11.84SRR523 pKa = 11.84NLSSDD528 pKa = 3.76IISNPYY534 pKa = 8.26IQKK537 pKa = 10.92ANN539 pKa = 3.44
MM1 pKa = 7.92RR2 pKa = 11.84VRR4 pKa = 11.84PLIIILVLLVLLWLNILPVIGLDD27 pKa = 3.34NSKK30 pKa = 9.92IAQAGIISAQEE41 pKa = 3.78YY42 pKa = 9.53AVNVYY47 pKa = 9.97SQSNEE52 pKa = 3.37AYY54 pKa = 8.7IALRR58 pKa = 11.84TVPYY62 pKa = 10.09IPPHH66 pKa = 6.01NLSCFQDD73 pKa = 4.53LINTYY78 pKa = 8.44NTTIQNIFSPIQDD91 pKa = 3.85QITSITSASTLPSSRR106 pKa = 11.84FAGLVVGAIALGVATSAQITAAVALTKK133 pKa = 10.39AQQNAQEE140 pKa = 4.53IIRR143 pKa = 11.84LRR145 pKa = 11.84DD146 pKa = 3.66SIQNTINAVNDD157 pKa = 3.24ITVGLSSIGVALSKK171 pKa = 10.05VQNYY175 pKa = 10.03LNDD178 pKa = 3.87VINPALQNLSCQVSALNLGIQLNLYY203 pKa = 8.18LTEE206 pKa = 3.8ITTIFGPQITNPSLTPLSIQALYY229 pKa = 9.57TLAGDD234 pKa = 4.04NLMQFLTRR242 pKa = 11.84YY243 pKa = 9.77GYY245 pKa = 11.41GEE247 pKa = 4.2TSVSSILEE255 pKa = 3.98SGLISAQIVSFDD267 pKa = 3.67KK268 pKa = 10.22QTGIAILYY276 pKa = 6.9VTLPSIATLSGSRR289 pKa = 11.84VTKK292 pKa = 10.54LMSVSVQTGVGEE304 pKa = 4.46GSAIVPSYY312 pKa = 10.7VIQQGTVIEE321 pKa = 4.24EE322 pKa = 4.64FIPDD326 pKa = 3.27SCIFTRR332 pKa = 11.84SDD334 pKa = 3.71VYY336 pKa = 10.36CTQLYY341 pKa = 10.78SKK343 pKa = 10.23LLPDD347 pKa = 5.99SILQCLQGSMADD359 pKa = 3.72CQFTRR364 pKa = 11.84SLGSFANRR372 pKa = 11.84FMTVAGGVIANCQTVLCRR390 pKa = 11.84CYY392 pKa = 10.24NPVMIIPQNNGIAVTLIDD410 pKa = 4.55GSLCKK415 pKa = 10.08EE416 pKa = 4.31LEE418 pKa = 4.17LEE420 pKa = 4.73GIRR423 pKa = 11.84LTMADD428 pKa = 4.18PVFASYY434 pKa = 11.11SRR436 pKa = 11.84DD437 pKa = 3.32LIINGNQFAPSDD449 pKa = 3.72ALDD452 pKa = 3.42ISSEE456 pKa = 4.02LGQLNNSISSATDD469 pKa = 3.24NLQKK473 pKa = 10.82AQEE476 pKa = 4.31SLNKK480 pKa = 10.31SIIPAATSSWLIILLFVLVSISLVIGCISIYY511 pKa = 10.44FIYY514 pKa = 10.23KK515 pKa = 9.93HH516 pKa = 5.42STTNRR521 pKa = 11.84SRR523 pKa = 11.84NLSSDD528 pKa = 3.76IISNPYY534 pKa = 8.26IQKK537 pKa = 10.92ANN539 pKa = 3.44
Molecular weight: 58.11 kDa
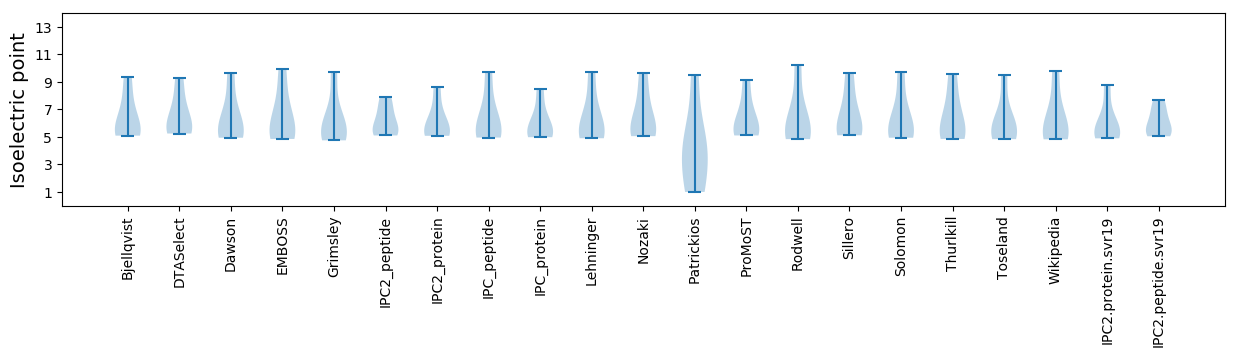
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6FGY9|C6FGY9_9MONO Fusion glycoprotein F0 OS=Avian metaavulavirus 7 OX=2560317 GN=F PE=3 SV=1
MM1 pKa = 7.55EE2 pKa = 5.78SISLGLYY9 pKa = 9.82VDD11 pKa = 4.54EE12 pKa = 5.46SDD14 pKa = 4.18PACSLLAFPIIMQTTSEE31 pKa = 4.29GKK33 pKa = 10.02KK34 pKa = 9.74VLQPQVRR41 pKa = 11.84INRR44 pKa = 11.84LGSISIEE51 pKa = 3.93GVRR54 pKa = 11.84AMFINTYY61 pKa = 10.31GFIEE65 pKa = 4.12EE66 pKa = 4.75RR67 pKa = 11.84PTEE70 pKa = 3.96RR71 pKa = 11.84TGFFQPGEE79 pKa = 4.15KK80 pKa = 9.96NQQQVVTAGMLTLGQIRR97 pKa = 11.84TNIDD101 pKa = 2.97PDD103 pKa = 4.21EE104 pKa = 5.55IGEE107 pKa = 4.14ACLRR111 pKa = 11.84LKK113 pKa = 11.14VNAKK117 pKa = 9.85KK118 pKa = 10.48SAASEE123 pKa = 4.05EE124 pKa = 4.43KK125 pKa = 9.97IVFSILEE132 pKa = 4.22KK133 pKa = 10.69PPALMTAPVVQDD145 pKa = 3.17GGLIAKK151 pKa = 10.21AEE153 pKa = 4.14GSIKK157 pKa = 10.68CPGKK161 pKa = 9.95MMSEE165 pKa = 3.29IHH167 pKa = 5.82YY168 pKa = 9.01SFRR171 pKa = 11.84VMFVSITMLDD181 pKa = 3.48NQSLYY186 pKa = 10.4RR187 pKa = 11.84VPTAISSFKK196 pKa = 11.01NKK198 pKa = 10.1ALYY201 pKa = 10.27SIQLEE206 pKa = 4.29VLLEE210 pKa = 4.03VDD212 pKa = 4.68VKK214 pKa = 11.12PEE216 pKa = 3.87SPQCKK221 pKa = 9.41FLADD225 pKa = 3.52QKK227 pKa = 9.68GKK229 pKa = 10.51KK230 pKa = 8.37VASVWFHH237 pKa = 6.27LCNSKK242 pKa = 8.46KK243 pKa = 9.2TNASGKK249 pKa = 8.92PRR251 pKa = 11.84SLEE254 pKa = 3.85DD255 pKa = 3.06MRR257 pKa = 11.84KK258 pKa = 9.26KK259 pKa = 10.5VRR261 pKa = 11.84DD262 pKa = 3.42MGIKK266 pKa = 10.28VSLADD271 pKa = 3.48LWGPTIIVRR280 pKa = 11.84ATGKK284 pKa = 8.8MSKK287 pKa = 10.81YY288 pKa = 9.09MLGFFSTSGTSCHH301 pKa = 6.37PVTKK305 pKa = 10.45SSPDD309 pKa = 3.17LAKK312 pKa = 10.48ILWSCSSTIIKK323 pKa = 10.42ANAIVQGSVKK333 pKa = 10.16VDD335 pKa = 3.22VLTLEE340 pKa = 5.2DD341 pKa = 3.68IQVSSAAKK349 pKa = 9.59INKK352 pKa = 9.67SGIGKK357 pKa = 9.13FNPFKK362 pKa = 10.81KK363 pKa = 10.52
MM1 pKa = 7.55EE2 pKa = 5.78SISLGLYY9 pKa = 9.82VDD11 pKa = 4.54EE12 pKa = 5.46SDD14 pKa = 4.18PACSLLAFPIIMQTTSEE31 pKa = 4.29GKK33 pKa = 10.02KK34 pKa = 9.74VLQPQVRR41 pKa = 11.84INRR44 pKa = 11.84LGSISIEE51 pKa = 3.93GVRR54 pKa = 11.84AMFINTYY61 pKa = 10.31GFIEE65 pKa = 4.12EE66 pKa = 4.75RR67 pKa = 11.84PTEE70 pKa = 3.96RR71 pKa = 11.84TGFFQPGEE79 pKa = 4.15KK80 pKa = 9.96NQQQVVTAGMLTLGQIRR97 pKa = 11.84TNIDD101 pKa = 2.97PDD103 pKa = 4.21EE104 pKa = 5.55IGEE107 pKa = 4.14ACLRR111 pKa = 11.84LKK113 pKa = 11.14VNAKK117 pKa = 9.85KK118 pKa = 10.48SAASEE123 pKa = 4.05EE124 pKa = 4.43KK125 pKa = 9.97IVFSILEE132 pKa = 4.22KK133 pKa = 10.69PPALMTAPVVQDD145 pKa = 3.17GGLIAKK151 pKa = 10.21AEE153 pKa = 4.14GSIKK157 pKa = 10.68CPGKK161 pKa = 9.95MMSEE165 pKa = 3.29IHH167 pKa = 5.82YY168 pKa = 9.01SFRR171 pKa = 11.84VMFVSITMLDD181 pKa = 3.48NQSLYY186 pKa = 10.4RR187 pKa = 11.84VPTAISSFKK196 pKa = 11.01NKK198 pKa = 10.1ALYY201 pKa = 10.27SIQLEE206 pKa = 4.29VLLEE210 pKa = 4.03VDD212 pKa = 4.68VKK214 pKa = 11.12PEE216 pKa = 3.87SPQCKK221 pKa = 9.41FLADD225 pKa = 3.52QKK227 pKa = 9.68GKK229 pKa = 10.51KK230 pKa = 8.37VASVWFHH237 pKa = 6.27LCNSKK242 pKa = 8.46KK243 pKa = 9.2TNASGKK249 pKa = 8.92PRR251 pKa = 11.84SLEE254 pKa = 3.85DD255 pKa = 3.06MRR257 pKa = 11.84KK258 pKa = 9.26KK259 pKa = 10.5VRR261 pKa = 11.84DD262 pKa = 3.42MGIKK266 pKa = 10.28VSLADD271 pKa = 3.48LWGPTIIVRR280 pKa = 11.84ATGKK284 pKa = 8.8MSKK287 pKa = 10.81YY288 pKa = 9.09MLGFFSTSGTSCHH301 pKa = 6.37PVTKK305 pKa = 10.45SSPDD309 pKa = 3.17LAKK312 pKa = 10.48ILWSCSSTIIKK323 pKa = 10.42ANAIVQGSVKK333 pKa = 10.16VDD335 pKa = 3.22VLTLEE340 pKa = 5.2DD341 pKa = 3.68IQVSSAAKK349 pKa = 9.59INKK352 pKa = 9.67SGIGKK357 pKa = 9.13FNPFKK362 pKa = 10.81KK363 pKa = 10.52
Molecular weight: 39.81 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4555 |
363 |
2227 |
759.2 |
84.23 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.828 ± 0.742 | 1.932 ± 0.36 |
4.83 ± 0.413 | 4.632 ± 0.416 |
3.403 ± 0.435 | 5.423 ± 0.383 |
1.361 ± 0.292 | 8.518 ± 0.793 |
5.005 ± 0.836 | 11.416 ± 0.725 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.261 ± 0.353 | 5.049 ± 0.486 |
4.479 ± 0.386 | 5.357 ± 0.619 |
4.193 ± 0.537 | 8.957 ± 0.603 |
6.213 ± 0.433 | 5.884 ± 0.393 |
1.076 ± 0.237 | 3.183 ± 0.505 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
