
Calanoida sp. copepod associated circular virus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.49
Get precalculated fractions of proteins
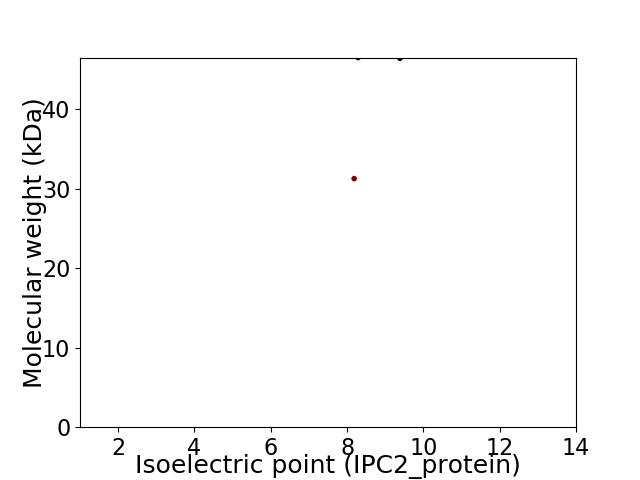
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0K1RLR5|A0A0K1RLR5_9CIRC ATP-dependent helicase Rep OS=Calanoida sp. copepod associated circular virus OX=1692244 PE=3 SV=1
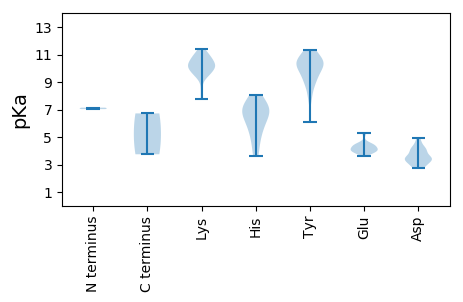
MM1 pKa = 7.14VNSGKK6 pKa = 9.56HH7 pKa = 3.65WCFTLNNYY15 pKa = 7.84TPEE18 pKa = 4.19EE19 pKa = 4.01EE20 pKa = 4.5SSIKK24 pKa = 10.72LVDD27 pKa = 3.49AVYY30 pKa = 10.32LVYY33 pKa = 10.62GRR35 pKa = 11.84EE36 pKa = 4.12RR37 pKa = 11.84GEE39 pKa = 3.78SDD41 pKa = 3.35TPHH44 pKa = 6.29LQGFVSFDD52 pKa = 2.88KK53 pKa = 10.81RR54 pKa = 11.84RR55 pKa = 11.84TLRR58 pKa = 11.84QVKK61 pKa = 9.43EE62 pKa = 3.88LLSDD66 pKa = 4.03RR67 pKa = 11.84VHH69 pKa = 7.01LEE71 pKa = 3.92SAKK74 pKa = 9.39GTPRR78 pKa = 11.84QASEE82 pKa = 4.16YY83 pKa = 9.38CKK85 pKa = 10.53KK86 pKa = 10.83DD87 pKa = 2.75GDD89 pKa = 4.15YY90 pKa = 11.21YY91 pKa = 11.11EE92 pKa = 5.27VGILPGGRR100 pKa = 11.84GTRR103 pKa = 11.84TDD105 pKa = 3.59LEE107 pKa = 4.56SVRR110 pKa = 11.84EE111 pKa = 4.17LIKK114 pKa = 11.01SGGSIRR120 pKa = 11.84DD121 pKa = 3.37VFDD124 pKa = 2.88KK125 pKa = 11.15HH126 pKa = 7.14FGVALRR132 pKa = 11.84YY133 pKa = 9.54HH134 pKa = 7.13RR135 pKa = 11.84GISMAINLRR144 pKa = 11.84GDD146 pKa = 3.05KK147 pKa = 10.71RR148 pKa = 11.84EE149 pKa = 3.99WVVDD153 pKa = 3.64VQVYY157 pKa = 9.32WGDD160 pKa = 3.48TGSGKK165 pKa = 7.75TKK167 pKa = 10.37KK168 pKa = 10.21AWTEE172 pKa = 3.6APDD175 pKa = 4.27AYY177 pKa = 10.62VHH179 pKa = 7.35PGGQWFDD186 pKa = 3.91GYY188 pKa = 9.43TGQPDD193 pKa = 4.05VIFDD197 pKa = 4.11DD198 pKa = 4.93FSGSEE203 pKa = 4.02FKK205 pKa = 10.14LTYY208 pKa = 10.26LLKK211 pKa = 11.04LLDD214 pKa = 4.7RR215 pKa = 11.84YY216 pKa = 10.12PMQVPVKK223 pKa = 9.76GGFVNWVPKK232 pKa = 10.39RR233 pKa = 11.84IFITSNLDD241 pKa = 2.82PRR243 pKa = 11.84TWYY246 pKa = 10.35KK247 pKa = 10.97DD248 pKa = 3.34AIQEE252 pKa = 4.02HH253 pKa = 5.49QAALMRR259 pKa = 11.84RR260 pKa = 11.84ITKK263 pKa = 8.43ITHH266 pKa = 4.58FTIPFHH272 pKa = 6.72
MM1 pKa = 7.14VNSGKK6 pKa = 9.56HH7 pKa = 3.65WCFTLNNYY15 pKa = 7.84TPEE18 pKa = 4.19EE19 pKa = 4.01EE20 pKa = 4.5SSIKK24 pKa = 10.72LVDD27 pKa = 3.49AVYY30 pKa = 10.32LVYY33 pKa = 10.62GRR35 pKa = 11.84EE36 pKa = 4.12RR37 pKa = 11.84GEE39 pKa = 3.78SDD41 pKa = 3.35TPHH44 pKa = 6.29LQGFVSFDD52 pKa = 2.88KK53 pKa = 10.81RR54 pKa = 11.84RR55 pKa = 11.84TLRR58 pKa = 11.84QVKK61 pKa = 9.43EE62 pKa = 3.88LLSDD66 pKa = 4.03RR67 pKa = 11.84VHH69 pKa = 7.01LEE71 pKa = 3.92SAKK74 pKa = 9.39GTPRR78 pKa = 11.84QASEE82 pKa = 4.16YY83 pKa = 9.38CKK85 pKa = 10.53KK86 pKa = 10.83DD87 pKa = 2.75GDD89 pKa = 4.15YY90 pKa = 11.21YY91 pKa = 11.11EE92 pKa = 5.27VGILPGGRR100 pKa = 11.84GTRR103 pKa = 11.84TDD105 pKa = 3.59LEE107 pKa = 4.56SVRR110 pKa = 11.84EE111 pKa = 4.17LIKK114 pKa = 11.01SGGSIRR120 pKa = 11.84DD121 pKa = 3.37VFDD124 pKa = 2.88KK125 pKa = 11.15HH126 pKa = 7.14FGVALRR132 pKa = 11.84YY133 pKa = 9.54HH134 pKa = 7.13RR135 pKa = 11.84GISMAINLRR144 pKa = 11.84GDD146 pKa = 3.05KK147 pKa = 10.71RR148 pKa = 11.84EE149 pKa = 3.99WVVDD153 pKa = 3.64VQVYY157 pKa = 9.32WGDD160 pKa = 3.48TGSGKK165 pKa = 7.75TKK167 pKa = 10.37KK168 pKa = 10.21AWTEE172 pKa = 3.6APDD175 pKa = 4.27AYY177 pKa = 10.62VHH179 pKa = 7.35PGGQWFDD186 pKa = 3.91GYY188 pKa = 9.43TGQPDD193 pKa = 4.05VIFDD197 pKa = 4.11DD198 pKa = 4.93FSGSEE203 pKa = 4.02FKK205 pKa = 10.14LTYY208 pKa = 10.26LLKK211 pKa = 11.04LLDD214 pKa = 4.7RR215 pKa = 11.84YY216 pKa = 10.12PMQVPVKK223 pKa = 9.76GGFVNWVPKK232 pKa = 10.39RR233 pKa = 11.84IFITSNLDD241 pKa = 2.82PRR243 pKa = 11.84TWYY246 pKa = 10.35KK247 pKa = 10.97DD248 pKa = 3.34AIQEE252 pKa = 4.02HH253 pKa = 5.49QAALMRR259 pKa = 11.84RR260 pKa = 11.84ITKK263 pKa = 8.43ITHH266 pKa = 4.58FTIPFHH272 pKa = 6.72
Molecular weight: 31.26 kDa
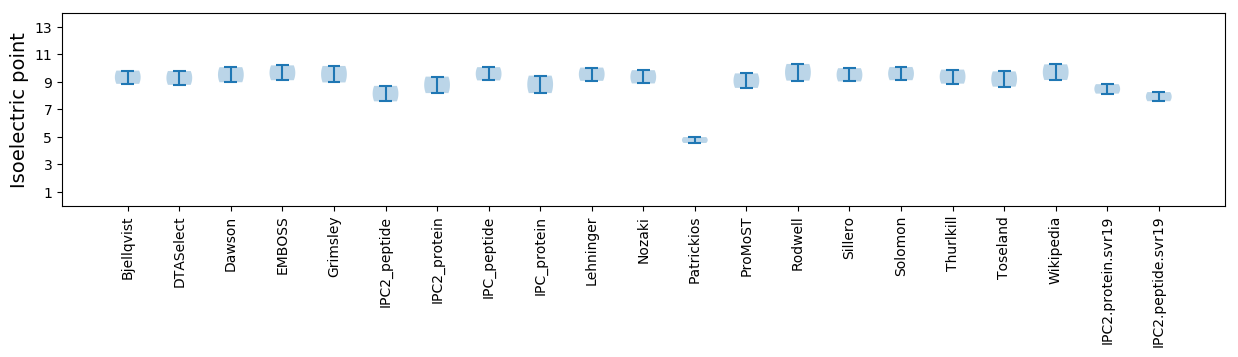
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0K1RLR5|A0A0K1RLR5_9CIRC ATP-dependent helicase Rep OS=Calanoida sp. copepod associated circular virus OX=1692244 PE=3 SV=1
MM1 pKa = 7.03VSSMTPYY8 pKa = 10.19SRR10 pKa = 11.84SRR12 pKa = 11.84YY13 pKa = 8.7GGVWLPAARR22 pKa = 11.84AAGRR26 pKa = 11.84LAIRR30 pKa = 11.84AGSRR34 pKa = 11.84YY35 pKa = 7.9VARR38 pKa = 11.84LLGKK42 pKa = 9.95RR43 pKa = 11.84KK44 pKa = 9.65RR45 pKa = 11.84ATTTAHH51 pKa = 5.47QPRR54 pKa = 11.84KK55 pKa = 9.52RR56 pKa = 11.84VYY58 pKa = 8.85TKK60 pKa = 9.1TKK62 pKa = 9.69RR63 pKa = 11.84YY64 pKa = 6.09MTGGYY69 pKa = 8.79IGRR72 pKa = 11.84RR73 pKa = 11.84VKK75 pKa = 10.13RR76 pKa = 11.84AKK78 pKa = 10.24RR79 pKa = 11.84RR80 pKa = 11.84TRR82 pKa = 11.84QNAATLYY89 pKa = 8.97GCSLKK94 pKa = 10.01TEE96 pKa = 4.43LGGVVTDD103 pKa = 4.05SKK105 pKa = 11.4AVYY108 pKa = 9.76VGHH111 pKa = 7.16CSVQPEE117 pKa = 5.26LILKK121 pKa = 9.65AAFRR125 pKa = 11.84AIVKK129 pKa = 10.01KK130 pKa = 10.68LYY132 pKa = 10.26DD133 pKa = 3.51GAGIRR138 pKa = 11.84IASWMDD144 pKa = 3.19KK145 pKa = 10.01PVNPAVLYY153 pKa = 10.5CKK155 pKa = 10.32LYY157 pKa = 10.15NQPASQSLNTLSYY170 pKa = 10.44TLLATDD176 pKa = 4.57TYY178 pKa = 11.35QDD180 pKa = 3.24IAEE183 pKa = 4.4GLLNVVVNYY192 pKa = 10.36INLTVAINTLPTFTYY207 pKa = 10.5FSLQHH212 pKa = 6.41GGTAVAEE219 pKa = 4.22IQAPQARR226 pKa = 11.84FYY228 pKa = 11.3VSTYY232 pKa = 9.62STLTVQNRR240 pKa = 11.84TLAGVSADD248 pKa = 3.44NDD250 pKa = 3.19EE251 pKa = 4.43STNVSKK257 pKa = 11.38NPVHH261 pKa = 6.55GKK263 pKa = 9.74LYY265 pKa = 10.23CGRR268 pKa = 11.84GNFFEE273 pKa = 4.49PSYY276 pKa = 10.79RR277 pKa = 11.84DD278 pKa = 3.45PNDD281 pKa = 3.38ISWVPLVCGRR291 pKa = 11.84TFGSMAATASGSLPLDD307 pKa = 3.35GMKK310 pKa = 10.35PPEE313 pKa = 3.92GSYY316 pKa = 10.89FSNCMSTSRR325 pKa = 11.84ITIKK329 pKa = 10.19PGEE332 pKa = 4.09IRR334 pKa = 11.84RR335 pKa = 11.84SVLKK339 pKa = 10.05YY340 pKa = 9.82KK341 pKa = 10.59RR342 pKa = 11.84SFQFTEE348 pKa = 4.35LFHH351 pKa = 8.06QLFNLIKK358 pKa = 10.09VYY360 pKa = 10.62SLSAQVPTMLGNMNMFGLEE379 pKa = 3.9KK380 pKa = 10.71LLDD383 pKa = 3.92SRR385 pKa = 11.84SDD387 pKa = 3.5EE388 pKa = 4.14PDD390 pKa = 2.84ISIGWEE396 pKa = 3.82VNNTVDD402 pKa = 4.23CGLSIKK408 pKa = 10.27RR409 pKa = 11.84CNPPAEE415 pKa = 4.42VIEE418 pKa = 4.41TT419 pKa = 3.75
MM1 pKa = 7.03VSSMTPYY8 pKa = 10.19SRR10 pKa = 11.84SRR12 pKa = 11.84YY13 pKa = 8.7GGVWLPAARR22 pKa = 11.84AAGRR26 pKa = 11.84LAIRR30 pKa = 11.84AGSRR34 pKa = 11.84YY35 pKa = 7.9VARR38 pKa = 11.84LLGKK42 pKa = 9.95RR43 pKa = 11.84KK44 pKa = 9.65RR45 pKa = 11.84ATTTAHH51 pKa = 5.47QPRR54 pKa = 11.84KK55 pKa = 9.52RR56 pKa = 11.84VYY58 pKa = 8.85TKK60 pKa = 9.1TKK62 pKa = 9.69RR63 pKa = 11.84YY64 pKa = 6.09MTGGYY69 pKa = 8.79IGRR72 pKa = 11.84RR73 pKa = 11.84VKK75 pKa = 10.13RR76 pKa = 11.84AKK78 pKa = 10.24RR79 pKa = 11.84RR80 pKa = 11.84TRR82 pKa = 11.84QNAATLYY89 pKa = 8.97GCSLKK94 pKa = 10.01TEE96 pKa = 4.43LGGVVTDD103 pKa = 4.05SKK105 pKa = 11.4AVYY108 pKa = 9.76VGHH111 pKa = 7.16CSVQPEE117 pKa = 5.26LILKK121 pKa = 9.65AAFRR125 pKa = 11.84AIVKK129 pKa = 10.01KK130 pKa = 10.68LYY132 pKa = 10.26DD133 pKa = 3.51GAGIRR138 pKa = 11.84IASWMDD144 pKa = 3.19KK145 pKa = 10.01PVNPAVLYY153 pKa = 10.5CKK155 pKa = 10.32LYY157 pKa = 10.15NQPASQSLNTLSYY170 pKa = 10.44TLLATDD176 pKa = 4.57TYY178 pKa = 11.35QDD180 pKa = 3.24IAEE183 pKa = 4.4GLLNVVVNYY192 pKa = 10.36INLTVAINTLPTFTYY207 pKa = 10.5FSLQHH212 pKa = 6.41GGTAVAEE219 pKa = 4.22IQAPQARR226 pKa = 11.84FYY228 pKa = 11.3VSTYY232 pKa = 9.62STLTVQNRR240 pKa = 11.84TLAGVSADD248 pKa = 3.44NDD250 pKa = 3.19EE251 pKa = 4.43STNVSKK257 pKa = 11.38NPVHH261 pKa = 6.55GKK263 pKa = 9.74LYY265 pKa = 10.23CGRR268 pKa = 11.84GNFFEE273 pKa = 4.49PSYY276 pKa = 10.79RR277 pKa = 11.84DD278 pKa = 3.45PNDD281 pKa = 3.38ISWVPLVCGRR291 pKa = 11.84TFGSMAATASGSLPLDD307 pKa = 3.35GMKK310 pKa = 10.35PPEE313 pKa = 3.92GSYY316 pKa = 10.89FSNCMSTSRR325 pKa = 11.84ITIKK329 pKa = 10.19PGEE332 pKa = 4.09IRR334 pKa = 11.84RR335 pKa = 11.84SVLKK339 pKa = 10.05YY340 pKa = 9.82KK341 pKa = 10.59RR342 pKa = 11.84SFQFTEE348 pKa = 4.35LFHH351 pKa = 8.06QLFNLIKK358 pKa = 10.09VYY360 pKa = 10.62SLSAQVPTMLGNMNMFGLEE379 pKa = 3.9KK380 pKa = 10.71LLDD383 pKa = 3.92SRR385 pKa = 11.84SDD387 pKa = 3.5EE388 pKa = 4.14PDD390 pKa = 2.84ISIGWEE396 pKa = 3.82VNNTVDD402 pKa = 4.23CGLSIKK408 pKa = 10.27RR409 pKa = 11.84CNPPAEE415 pKa = 4.42VIEE418 pKa = 4.41TT419 pKa = 3.75
Molecular weight: 46.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
691 |
272 |
419 |
345.5 |
38.82 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.512 ± 1.469 | 1.447 ± 0.424 |
4.92 ± 1.448 | 4.197 ± 0.784 |
3.763 ± 0.605 | 8.104 ± 0.647 |
2.026 ± 0.764 | 4.631 ± 0.088 |
6.078 ± 0.54 | 8.249 ± 0.533 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.026 ± 0.331 | 3.907 ± 1.013 |
4.776 ± 0.217 | 3.184 ± 0.074 |
7.381 ± 0.016 | 7.381 ± 0.892 |
7.236 ± 0.587 | 7.525 ± 0.116 |
1.592 ± 0.584 | 5.065 ± 0.17 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
