
Tibetan frog hepatitis B virus
Taxonomy: Viruses; Riboviria; Pararnavirae; Artverviricota; Revtraviricetes; Blubervirales; Hepadnaviridae; Herpetohepadnavirus
Average proteome isoelectric point is 8.36
Get precalculated fractions of proteins
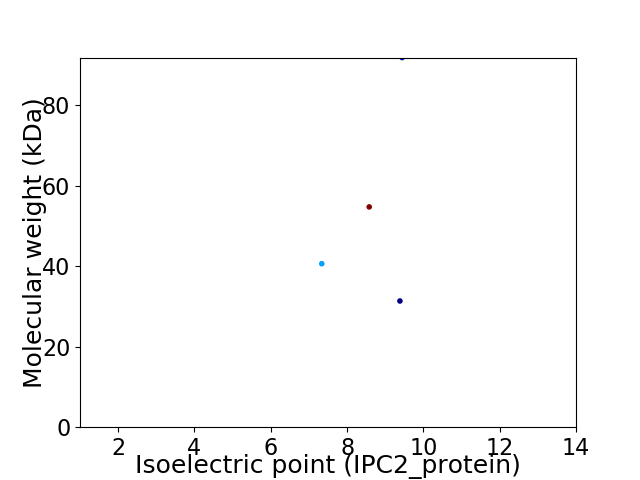
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|A0A193AUB7|A0A193AUB7_9HEPA Isoform of A0A193AU90 Large envelope protein OS=Tibetan frog hepatitis B virus OX=2169919 GN=PreS PE=4 SV=1
MM1 pKa = 7.74RR2 pKa = 11.84FFARR6 pKa = 11.84VWSWIGGNPSQHH18 pKa = 5.7VSAQEE23 pKa = 3.64HH24 pKa = 6.68RR25 pKa = 11.84DD26 pKa = 3.61RR27 pKa = 11.84MEE29 pKa = 4.8DD30 pKa = 3.05PHH32 pKa = 8.71AFQQYY37 pKa = 10.19LATLPDD43 pKa = 3.47INRR46 pKa = 11.84AALLSLNKK54 pKa = 9.91HH55 pKa = 5.56FRR57 pKa = 11.84LDD59 pKa = 3.37NHH61 pKa = 6.58QGFLQILDD69 pKa = 3.51NATFAEE75 pKa = 4.48IAEE78 pKa = 4.17HH79 pKa = 6.38YY80 pKa = 10.35YY81 pKa = 10.69RR82 pKa = 11.84FRR84 pKa = 11.84QAYY87 pKa = 8.85ANHH90 pKa = 6.94SITWTPPHH98 pKa = 6.42EE99 pKa = 4.03EE100 pKa = 4.5EE101 pKa = 4.36YY102 pKa = 10.18WEE104 pKa = 4.02EE105 pKa = 3.68HH106 pKa = 5.64ALYY109 pKa = 9.41PEE111 pKa = 4.21KK112 pKa = 10.45AAPVEE117 pKa = 4.22PIIPVPGPKK126 pKa = 9.33RR127 pKa = 11.84APPLKK132 pKa = 10.07PRR134 pKa = 11.84PQPRR138 pKa = 11.84NRR140 pKa = 11.84TKK142 pKa = 10.65LRR144 pKa = 11.84KK145 pKa = 9.1PSDD148 pKa = 4.59AIPVLLAIPTSSSTEE163 pKa = 3.99VGRR166 pKa = 11.84LPYY169 pKa = 10.08QIYY172 pKa = 9.16PHH174 pKa = 6.95EE175 pKa = 4.85LLPIPKK181 pKa = 9.7LEE183 pKa = 4.39EE184 pKa = 4.07KK185 pKa = 9.78STTANMSGAVSPALWFPVVALVVYY209 pKa = 9.41FLLTKK214 pKa = 10.47ARR216 pKa = 11.84EE217 pKa = 3.99ILAKK221 pKa = 10.3LDD223 pKa = 3.46WWWTSHH229 pKa = 5.78SFRR232 pKa = 11.84GANGEE237 pKa = 5.09CIFQDD242 pKa = 4.41TSPQTQKK249 pKa = 10.86HH250 pKa = 5.13LAGFCPLTCGGFLWTSLRR268 pKa = 11.84LFTICLIILLAVCGYY283 pKa = 10.13LFLMEE288 pKa = 5.5KK289 pKa = 10.7GFTSLGEE296 pKa = 4.08QLWEE300 pKa = 4.46LGSAPGTLLSSLLSSLKK317 pKa = 10.27GSGNIFLSIALLTWMTSSYY336 pKa = 10.02LTLTPATSLLLLTGLWATYY355 pKa = 11.26NNMGSGG361 pKa = 3.47
MM1 pKa = 7.74RR2 pKa = 11.84FFARR6 pKa = 11.84VWSWIGGNPSQHH18 pKa = 5.7VSAQEE23 pKa = 3.64HH24 pKa = 6.68RR25 pKa = 11.84DD26 pKa = 3.61RR27 pKa = 11.84MEE29 pKa = 4.8DD30 pKa = 3.05PHH32 pKa = 8.71AFQQYY37 pKa = 10.19LATLPDD43 pKa = 3.47INRR46 pKa = 11.84AALLSLNKK54 pKa = 9.91HH55 pKa = 5.56FRR57 pKa = 11.84LDD59 pKa = 3.37NHH61 pKa = 6.58QGFLQILDD69 pKa = 3.51NATFAEE75 pKa = 4.48IAEE78 pKa = 4.17HH79 pKa = 6.38YY80 pKa = 10.35YY81 pKa = 10.69RR82 pKa = 11.84FRR84 pKa = 11.84QAYY87 pKa = 8.85ANHH90 pKa = 6.94SITWTPPHH98 pKa = 6.42EE99 pKa = 4.03EE100 pKa = 4.5EE101 pKa = 4.36YY102 pKa = 10.18WEE104 pKa = 4.02EE105 pKa = 3.68HH106 pKa = 5.64ALYY109 pKa = 9.41PEE111 pKa = 4.21KK112 pKa = 10.45AAPVEE117 pKa = 4.22PIIPVPGPKK126 pKa = 9.33RR127 pKa = 11.84APPLKK132 pKa = 10.07PRR134 pKa = 11.84PQPRR138 pKa = 11.84NRR140 pKa = 11.84TKK142 pKa = 10.65LRR144 pKa = 11.84KK145 pKa = 9.1PSDD148 pKa = 4.59AIPVLLAIPTSSSTEE163 pKa = 3.99VGRR166 pKa = 11.84LPYY169 pKa = 10.08QIYY172 pKa = 9.16PHH174 pKa = 6.95EE175 pKa = 4.85LLPIPKK181 pKa = 9.7LEE183 pKa = 4.39EE184 pKa = 4.07KK185 pKa = 9.78STTANMSGAVSPALWFPVVALVVYY209 pKa = 9.41FLLTKK214 pKa = 10.47ARR216 pKa = 11.84EE217 pKa = 3.99ILAKK221 pKa = 10.3LDD223 pKa = 3.46WWWTSHH229 pKa = 5.78SFRR232 pKa = 11.84GANGEE237 pKa = 5.09CIFQDD242 pKa = 4.41TSPQTQKK249 pKa = 10.86HH250 pKa = 5.13LAGFCPLTCGGFLWTSLRR268 pKa = 11.84LFTICLIILLAVCGYY283 pKa = 10.13LFLMEE288 pKa = 5.5KK289 pKa = 10.7GFTSLGEE296 pKa = 4.08QLWEE300 pKa = 4.46LGSAPGTLLSSLLSSLKK317 pKa = 10.27GSGNIFLSIALLTWMTSSYY336 pKa = 10.02LTLTPATSLLLLTGLWATYY355 pKa = 11.26NNMGSGG361 pKa = 3.47
Molecular weight: 40.63 kDa
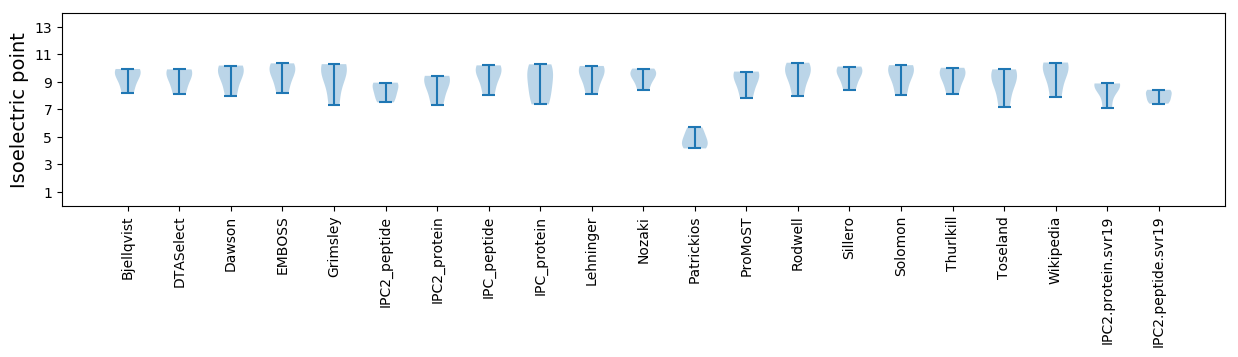
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A193AU90|A0A193AU90_9HEPA Large envelope protein OS=Tibetan frog hepatitis B virus OX=2169919 GN=PreS PE=4 SV=1
MM1 pKa = 7.27KK2 pKa = 10.31RR3 pKa = 11.84LWSRR7 pKa = 11.84NVRR10 pKa = 11.84GRR12 pKa = 11.84PGPILEE18 pKa = 4.34EE19 pKa = 4.21LEE21 pKa = 4.46GALTQAGTEE30 pKa = 4.13DD31 pKa = 4.15YY32 pKa = 10.4IIRR35 pKa = 11.84GQSGDD40 pKa = 4.54LIVQGKK46 pKa = 9.34EE47 pKa = 3.87DD48 pKa = 4.41LDD50 pKa = 4.55GGVLPLQAEE59 pKa = 4.63PEE61 pKa = 4.18RR62 pKa = 11.84LLGPLKK68 pKa = 10.63GLYY71 pKa = 10.17QNDD74 pKa = 3.68VPPFNPSWEE83 pKa = 4.1IPTIQGVHH91 pKa = 5.28LQEE94 pKa = 5.24DD95 pKa = 4.99IIRR98 pKa = 11.84EE99 pKa = 3.96IPDD102 pKa = 3.24RR103 pKa = 11.84RR104 pKa = 11.84WAYY107 pKa = 9.67IYY109 pKa = 9.53PIRR112 pKa = 11.84FWPNEE117 pKa = 3.6VKK119 pKa = 10.52YY120 pKa = 10.4RR121 pKa = 11.84PRR123 pKa = 11.84EE124 pKa = 3.6QGITDD129 pKa = 4.41KK130 pKa = 11.33YY131 pKa = 9.37PEE133 pKa = 3.98HH134 pKa = 6.21TRR136 pKa = 11.84AHH138 pKa = 5.52SEE140 pKa = 4.08KK141 pKa = 10.57VSLYY145 pKa = 10.91LQTLYY150 pKa = 11.51NNGILYY156 pKa = 10.03KK157 pKa = 10.21RR158 pKa = 11.84EE159 pKa = 3.99SRR161 pKa = 11.84HH162 pKa = 5.58VLLFNDD168 pKa = 4.58NPYY171 pKa = 10.65SWEE174 pKa = 4.28SNNLLPCHH182 pKa = 5.69CHH184 pKa = 6.97KK185 pKa = 11.11SQDD188 pKa = 3.74YY189 pKa = 10.29EE190 pKa = 4.29GLGRR194 pKa = 11.84QPPSRR199 pKa = 11.84RR200 pKa = 11.84SNEE203 pKa = 3.58VLRR206 pKa = 11.84PSVEE210 pKa = 3.88LDD212 pKa = 2.73RR213 pKa = 11.84GKK215 pKa = 9.97PLTTRR220 pKa = 11.84LSPRR224 pKa = 11.84AQRR227 pKa = 11.84PDD229 pKa = 3.01GRR231 pKa = 11.84PSRR234 pKa = 11.84FPAILSNPAGHH245 pKa = 5.97QPSGIVKK252 pKa = 9.52FKK254 pKa = 9.42QTLPAGQSPRR264 pKa = 11.84ISTDD268 pKa = 2.34TRR270 pKa = 11.84QCHH273 pKa = 4.33VRR275 pKa = 11.84GNSRR279 pKa = 11.84TLLQIPPSICQPQHH293 pKa = 6.93NMDD296 pKa = 3.52SSPRR300 pKa = 11.84GGILGGTRR308 pKa = 11.84PLSRR312 pKa = 11.84EE313 pKa = 4.0GGSGRR318 pKa = 11.84TYY320 pKa = 9.53HH321 pKa = 6.15TGSRR325 pKa = 11.84AQKK328 pKa = 9.91SSSSKK333 pKa = 9.64ATSSTPKK340 pKa = 9.6QDD342 pKa = 3.38KK343 pKa = 9.65AQEE346 pKa = 4.19AIRR349 pKa = 11.84RR350 pKa = 11.84HH351 pKa = 4.41TCAARR356 pKa = 11.84NSYY359 pKa = 9.12QQLNRR364 pKa = 11.84SRR366 pKa = 11.84PASISNLPPRR376 pKa = 11.84VTTYY380 pKa = 10.67PEE382 pKa = 4.15AGRR385 pKa = 11.84KK386 pKa = 8.63EE387 pKa = 4.23YY388 pKa = 10.55NCKK391 pKa = 10.1HH392 pKa = 5.59EE393 pKa = 4.28RR394 pKa = 11.84RR395 pKa = 11.84CFSRR399 pKa = 11.84SVVPSGCTGRR409 pKa = 11.84VFLVNKK415 pKa = 10.37SSGNSSQARR424 pKa = 11.84LVVDD428 pKa = 4.9FSQFSRR434 pKa = 11.84GKK436 pKa = 10.11RR437 pKa = 11.84RR438 pKa = 11.84VHH440 pKa = 6.11FPRR443 pKa = 11.84YY444 pKa = 9.23FSPNTEE450 pKa = 3.89ALSRR454 pKa = 11.84ILPPHH459 pKa = 6.05MWRR462 pKa = 11.84ISLDD466 pKa = 3.43LAEE469 pKa = 5.16AFYY472 pKa = 10.67HH473 pKa = 6.54LPYY476 pKa = 9.06HH477 pKa = 6.3TSSSLRR483 pKa = 11.84LSVSDD488 pKa = 3.54GKK490 pKa = 10.46RR491 pKa = 11.84VYY493 pKa = 10.7LFRR496 pKa = 11.84RR497 pKa = 11.84AAMGVGLSPWNLTLFTSIIAEE518 pKa = 4.52RR519 pKa = 11.84IRR521 pKa = 11.84KK522 pKa = 8.41YY523 pKa = 10.97FSIHH527 pKa = 5.81CFAYY531 pKa = 9.43MDD533 pKa = 4.75DD534 pKa = 5.59FILSHH539 pKa = 6.66PNPRR543 pKa = 11.84YY544 pKa = 10.28LIAAANGVVGYY555 pKa = 8.56LQQYY559 pKa = 7.3GVRR562 pKa = 11.84INFDD566 pKa = 3.45KK567 pKa = 10.8STLTPTRR574 pKa = 11.84QIVFMGLQITPTHH587 pKa = 5.01IHH589 pKa = 4.06VTKK592 pKa = 10.68EE593 pKa = 3.88KK594 pKa = 10.15FQKK597 pKa = 10.65LHH599 pKa = 7.25DD600 pKa = 4.91LLTSINTDD608 pKa = 2.48VFYY611 pKa = 10.73DD612 pKa = 3.41WKK614 pKa = 10.37ILQRR618 pKa = 11.84FAGHH622 pKa = 6.6FNQIVAFSLFGSHH635 pKa = 6.63ILGPIYY641 pKa = 10.48LAITNKK647 pKa = 10.35ADD649 pKa = 3.34FCFTKK654 pKa = 10.41SYY656 pKa = 9.54VTDD659 pKa = 3.56LRR661 pKa = 11.84YY662 pKa = 10.34VLNHH666 pKa = 6.19VYY668 pKa = 10.71DD669 pKa = 3.84LRR671 pKa = 11.84LRR673 pKa = 11.84ASPKK677 pKa = 10.22HH678 pKa = 5.6IVPSVAADD686 pKa = 3.24ATLTHH691 pKa = 6.63GGIATRR697 pKa = 11.84QGEE700 pKa = 4.57EE701 pKa = 3.59IQLTFSEE708 pKa = 4.24QRR710 pKa = 11.84PVHH713 pKa = 4.89VQEE716 pKa = 4.8LLIALVALLCFVPKK730 pKa = 10.69GLLSDD735 pKa = 3.72STFVVHH741 pKa = 6.6KK742 pKa = 10.13QLHH745 pKa = 4.94TLPYY749 pKa = 8.59ALAAVGHH756 pKa = 5.83YY757 pKa = 10.1LLSRR761 pKa = 11.84AKK763 pKa = 8.45VTYY766 pKa = 10.41VHH768 pKa = 6.77TSLNPADD775 pKa = 3.8PVSRR779 pKa = 11.84LLANHH784 pKa = 6.53SAPRR788 pKa = 11.84WPLLVHH794 pKa = 6.22TSLVKK799 pKa = 10.04PLYY802 pKa = 10.17IPWSLLRR809 pKa = 11.84SPP811 pKa = 4.9
MM1 pKa = 7.27KK2 pKa = 10.31RR3 pKa = 11.84LWSRR7 pKa = 11.84NVRR10 pKa = 11.84GRR12 pKa = 11.84PGPILEE18 pKa = 4.34EE19 pKa = 4.21LEE21 pKa = 4.46GALTQAGTEE30 pKa = 4.13DD31 pKa = 4.15YY32 pKa = 10.4IIRR35 pKa = 11.84GQSGDD40 pKa = 4.54LIVQGKK46 pKa = 9.34EE47 pKa = 3.87DD48 pKa = 4.41LDD50 pKa = 4.55GGVLPLQAEE59 pKa = 4.63PEE61 pKa = 4.18RR62 pKa = 11.84LLGPLKK68 pKa = 10.63GLYY71 pKa = 10.17QNDD74 pKa = 3.68VPPFNPSWEE83 pKa = 4.1IPTIQGVHH91 pKa = 5.28LQEE94 pKa = 5.24DD95 pKa = 4.99IIRR98 pKa = 11.84EE99 pKa = 3.96IPDD102 pKa = 3.24RR103 pKa = 11.84RR104 pKa = 11.84WAYY107 pKa = 9.67IYY109 pKa = 9.53PIRR112 pKa = 11.84FWPNEE117 pKa = 3.6VKK119 pKa = 10.52YY120 pKa = 10.4RR121 pKa = 11.84PRR123 pKa = 11.84EE124 pKa = 3.6QGITDD129 pKa = 4.41KK130 pKa = 11.33YY131 pKa = 9.37PEE133 pKa = 3.98HH134 pKa = 6.21TRR136 pKa = 11.84AHH138 pKa = 5.52SEE140 pKa = 4.08KK141 pKa = 10.57VSLYY145 pKa = 10.91LQTLYY150 pKa = 11.51NNGILYY156 pKa = 10.03KK157 pKa = 10.21RR158 pKa = 11.84EE159 pKa = 3.99SRR161 pKa = 11.84HH162 pKa = 5.58VLLFNDD168 pKa = 4.58NPYY171 pKa = 10.65SWEE174 pKa = 4.28SNNLLPCHH182 pKa = 5.69CHH184 pKa = 6.97KK185 pKa = 11.11SQDD188 pKa = 3.74YY189 pKa = 10.29EE190 pKa = 4.29GLGRR194 pKa = 11.84QPPSRR199 pKa = 11.84RR200 pKa = 11.84SNEE203 pKa = 3.58VLRR206 pKa = 11.84PSVEE210 pKa = 3.88LDD212 pKa = 2.73RR213 pKa = 11.84GKK215 pKa = 9.97PLTTRR220 pKa = 11.84LSPRR224 pKa = 11.84AQRR227 pKa = 11.84PDD229 pKa = 3.01GRR231 pKa = 11.84PSRR234 pKa = 11.84FPAILSNPAGHH245 pKa = 5.97QPSGIVKK252 pKa = 9.52FKK254 pKa = 9.42QTLPAGQSPRR264 pKa = 11.84ISTDD268 pKa = 2.34TRR270 pKa = 11.84QCHH273 pKa = 4.33VRR275 pKa = 11.84GNSRR279 pKa = 11.84TLLQIPPSICQPQHH293 pKa = 6.93NMDD296 pKa = 3.52SSPRR300 pKa = 11.84GGILGGTRR308 pKa = 11.84PLSRR312 pKa = 11.84EE313 pKa = 4.0GGSGRR318 pKa = 11.84TYY320 pKa = 9.53HH321 pKa = 6.15TGSRR325 pKa = 11.84AQKK328 pKa = 9.91SSSSKK333 pKa = 9.64ATSSTPKK340 pKa = 9.6QDD342 pKa = 3.38KK343 pKa = 9.65AQEE346 pKa = 4.19AIRR349 pKa = 11.84RR350 pKa = 11.84HH351 pKa = 4.41TCAARR356 pKa = 11.84NSYY359 pKa = 9.12QQLNRR364 pKa = 11.84SRR366 pKa = 11.84PASISNLPPRR376 pKa = 11.84VTTYY380 pKa = 10.67PEE382 pKa = 4.15AGRR385 pKa = 11.84KK386 pKa = 8.63EE387 pKa = 4.23YY388 pKa = 10.55NCKK391 pKa = 10.1HH392 pKa = 5.59EE393 pKa = 4.28RR394 pKa = 11.84RR395 pKa = 11.84CFSRR399 pKa = 11.84SVVPSGCTGRR409 pKa = 11.84VFLVNKK415 pKa = 10.37SSGNSSQARR424 pKa = 11.84LVVDD428 pKa = 4.9FSQFSRR434 pKa = 11.84GKK436 pKa = 10.11RR437 pKa = 11.84RR438 pKa = 11.84VHH440 pKa = 6.11FPRR443 pKa = 11.84YY444 pKa = 9.23FSPNTEE450 pKa = 3.89ALSRR454 pKa = 11.84ILPPHH459 pKa = 6.05MWRR462 pKa = 11.84ISLDD466 pKa = 3.43LAEE469 pKa = 5.16AFYY472 pKa = 10.67HH473 pKa = 6.54LPYY476 pKa = 9.06HH477 pKa = 6.3TSSSLRR483 pKa = 11.84LSVSDD488 pKa = 3.54GKK490 pKa = 10.46RR491 pKa = 11.84VYY493 pKa = 10.7LFRR496 pKa = 11.84RR497 pKa = 11.84AAMGVGLSPWNLTLFTSIIAEE518 pKa = 4.52RR519 pKa = 11.84IRR521 pKa = 11.84KK522 pKa = 8.41YY523 pKa = 10.97FSIHH527 pKa = 5.81CFAYY531 pKa = 9.43MDD533 pKa = 4.75DD534 pKa = 5.59FILSHH539 pKa = 6.66PNPRR543 pKa = 11.84YY544 pKa = 10.28LIAAANGVVGYY555 pKa = 8.56LQQYY559 pKa = 7.3GVRR562 pKa = 11.84INFDD566 pKa = 3.45KK567 pKa = 10.8STLTPTRR574 pKa = 11.84QIVFMGLQITPTHH587 pKa = 5.01IHH589 pKa = 4.06VTKK592 pKa = 10.68EE593 pKa = 3.88KK594 pKa = 10.15FQKK597 pKa = 10.65LHH599 pKa = 7.25DD600 pKa = 4.91LLTSINTDD608 pKa = 2.48VFYY611 pKa = 10.73DD612 pKa = 3.41WKK614 pKa = 10.37ILQRR618 pKa = 11.84FAGHH622 pKa = 6.6FNQIVAFSLFGSHH635 pKa = 6.63ILGPIYY641 pKa = 10.48LAITNKK647 pKa = 10.35ADD649 pKa = 3.34FCFTKK654 pKa = 10.41SYY656 pKa = 9.54VTDD659 pKa = 3.56LRR661 pKa = 11.84YY662 pKa = 10.34VLNHH666 pKa = 6.19VYY668 pKa = 10.71DD669 pKa = 3.84LRR671 pKa = 11.84LRR673 pKa = 11.84ASPKK677 pKa = 10.22HH678 pKa = 5.6IVPSVAADD686 pKa = 3.24ATLTHH691 pKa = 6.63GGIATRR697 pKa = 11.84QGEE700 pKa = 4.57EE701 pKa = 3.59IQLTFSEE708 pKa = 4.24QRR710 pKa = 11.84PVHH713 pKa = 4.89VQEE716 pKa = 4.8LLIALVALLCFVPKK730 pKa = 10.69GLLSDD735 pKa = 3.72STFVVHH741 pKa = 6.6KK742 pKa = 10.13QLHH745 pKa = 4.94TLPYY749 pKa = 8.59ALAAVGHH756 pKa = 5.83YY757 pKa = 10.1LLSRR761 pKa = 11.84AKK763 pKa = 8.45VTYY766 pKa = 10.41VHH768 pKa = 6.77TSLNPADD775 pKa = 3.8PVSRR779 pKa = 11.84LLANHH784 pKa = 6.53SAPRR788 pKa = 11.84WPLLVHH794 pKa = 6.22TSLVKK799 pKa = 10.04PLYY802 pKa = 10.17IPWSLLRR809 pKa = 11.84SPP811 pKa = 4.9
Molecular weight: 91.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
1928 |
266 |
811 |
482.0 |
54.65 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.587 ± 0.679 | 1.297 ± 0.215 |
3.06 ± 0.472 | 4.824 ± 0.496 |
4.253 ± 0.301 | 6.172 ± 0.302 |
3.683 ± 0.262 | 5.394 ± 0.293 |
3.631 ± 0.578 | 11.463 ± 0.736 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.297 ± 0.317 | 3.786 ± 0.256 |
7.988 ± 0.253 | 4.253 ± 0.371 |
7.573 ± 1.401 | 8.039 ± 0.626 |
6.38 ± 0.618 | 4.409 ± 0.833 |
2.127 ± 0.442 | 3.786 ± 0.181 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
