
Aurantiacibacter spongiae
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Erythrobacteraceae; Aurantiacibacter
Average proteome isoelectric point is 6.07
Get precalculated fractions of proteins
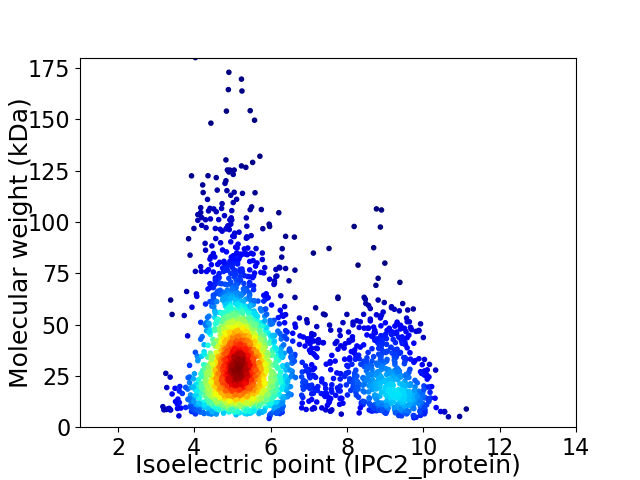
Virtual 2D-PAGE plot for 2690 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A3N5CPX8|A0A3N5CPX8_9SPHN Thiolase family protein OS=Aurantiacibacter spongiae OX=2488860 GN=EG799_02660 PE=3 SV=1
MM1 pKa = 7.33TNISRR6 pKa = 11.84NLILGCSLLALSACGPDD23 pKa = 4.44EE24 pKa = 4.08IASPGTGGDD33 pKa = 3.45IIINNPSPTPSPTPTPTPTSGGVTAAAGCPTIADD67 pKa = 3.79AQGLTDD73 pKa = 4.27EE74 pKa = 4.96GTISGPTGEE83 pKa = 4.53YY84 pKa = 9.35RR85 pKa = 11.84VCATPIRR92 pKa = 11.84FNSSSTLSYY101 pKa = 11.3VPGLLYY107 pKa = 10.51RR108 pKa = 11.84LPGQVSVGTDD118 pKa = 3.56GGPAPDD124 pKa = 4.25NSDD127 pKa = 3.54GFDD130 pKa = 3.59DD131 pKa = 4.16TDD133 pKa = 3.66VEE135 pKa = 4.4LTIEE139 pKa = 4.37PGVIIYY145 pKa = 10.64ASGSSFLNVTRR156 pKa = 11.84GNSIDD161 pKa = 3.75ANGTEE166 pKa = 4.41DD167 pKa = 3.79RR168 pKa = 11.84PIIFTSRR175 pKa = 11.84DD176 pKa = 3.4NILGLNNDD184 pKa = 3.64NSSGQWGGVVLSGRR198 pKa = 11.84APVTDD203 pKa = 4.52CFQPGATPGTVNCEE217 pKa = 4.05RR218 pKa = 11.84QVEE221 pKa = 4.6GAADD225 pKa = 3.69PNFFGGATPDD235 pKa = 3.85DD236 pKa = 4.42SSGSMSYY243 pKa = 10.01VQIRR247 pKa = 11.84FSGFVLSGDD256 pKa = 4.13NEE258 pKa = 4.34LQSLTTGGVGSGTDD272 pKa = 3.18LSYY275 pKa = 10.78IQSVNSSDD283 pKa = 4.32DD284 pKa = 3.38GAEE287 pKa = 4.02FFGGRR292 pKa = 11.84VNLSHH297 pKa = 7.28LIVAGAEE304 pKa = 3.94DD305 pKa = 5.08DD306 pKa = 5.31SLDD309 pKa = 3.36TDD311 pKa = 3.83TGVKK315 pKa = 10.69ANFQYY320 pKa = 11.2VLAAQRR326 pKa = 11.84SGVGDD331 pKa = 3.51TMIEE335 pKa = 3.9ADD337 pKa = 3.88SSNGLEE343 pKa = 4.42DD344 pKa = 3.14QTPRR348 pKa = 11.84QNTQVANFTFIQRR361 pKa = 11.84RR362 pKa = 11.84SGDD365 pKa = 3.2QVIRR369 pKa = 11.84LRR371 pKa = 11.84GGTDD375 pKa = 2.93YY376 pKa = 11.6GLYY379 pKa = 9.99NGVVIDD385 pKa = 4.1QSSSGTPCIRR395 pKa = 11.84IDD397 pKa = 3.6DD398 pKa = 4.08QEE400 pKa = 4.46TIRR403 pKa = 11.84AANAGLDD410 pKa = 3.76DD411 pKa = 4.03VGPPRR416 pKa = 11.84FNSVALDD423 pKa = 3.52CQTNFRR429 pKa = 11.84AGSDD433 pKa = 3.58GVTVADD439 pKa = 3.91VEE441 pKa = 4.92AILTSGTGNTLDD453 pKa = 4.16YY454 pKa = 11.4DD455 pKa = 3.74NTIQNGYY462 pKa = 10.3LNGANEE468 pKa = 4.68DD469 pKa = 3.71GYY471 pKa = 11.12MPIFDD476 pKa = 4.02VTSLSSFFEE485 pKa = 4.18SASFIGAVSASNDD498 pKa = 2.81WTQGWTCDD506 pKa = 3.18SAAITFGNNTGNCLTLPVYY525 pKa = 10.6
MM1 pKa = 7.33TNISRR6 pKa = 11.84NLILGCSLLALSACGPDD23 pKa = 4.44EE24 pKa = 4.08IASPGTGGDD33 pKa = 3.45IIINNPSPTPSPTPTPTPTSGGVTAAAGCPTIADD67 pKa = 3.79AQGLTDD73 pKa = 4.27EE74 pKa = 4.96GTISGPTGEE83 pKa = 4.53YY84 pKa = 9.35RR85 pKa = 11.84VCATPIRR92 pKa = 11.84FNSSSTLSYY101 pKa = 11.3VPGLLYY107 pKa = 10.51RR108 pKa = 11.84LPGQVSVGTDD118 pKa = 3.56GGPAPDD124 pKa = 4.25NSDD127 pKa = 3.54GFDD130 pKa = 3.59DD131 pKa = 4.16TDD133 pKa = 3.66VEE135 pKa = 4.4LTIEE139 pKa = 4.37PGVIIYY145 pKa = 10.64ASGSSFLNVTRR156 pKa = 11.84GNSIDD161 pKa = 3.75ANGTEE166 pKa = 4.41DD167 pKa = 3.79RR168 pKa = 11.84PIIFTSRR175 pKa = 11.84DD176 pKa = 3.4NILGLNNDD184 pKa = 3.64NSSGQWGGVVLSGRR198 pKa = 11.84APVTDD203 pKa = 4.52CFQPGATPGTVNCEE217 pKa = 4.05RR218 pKa = 11.84QVEE221 pKa = 4.6GAADD225 pKa = 3.69PNFFGGATPDD235 pKa = 3.85DD236 pKa = 4.42SSGSMSYY243 pKa = 10.01VQIRR247 pKa = 11.84FSGFVLSGDD256 pKa = 4.13NEE258 pKa = 4.34LQSLTTGGVGSGTDD272 pKa = 3.18LSYY275 pKa = 10.78IQSVNSSDD283 pKa = 4.32DD284 pKa = 3.38GAEE287 pKa = 4.02FFGGRR292 pKa = 11.84VNLSHH297 pKa = 7.28LIVAGAEE304 pKa = 3.94DD305 pKa = 5.08DD306 pKa = 5.31SLDD309 pKa = 3.36TDD311 pKa = 3.83TGVKK315 pKa = 10.69ANFQYY320 pKa = 11.2VLAAQRR326 pKa = 11.84SGVGDD331 pKa = 3.51TMIEE335 pKa = 3.9ADD337 pKa = 3.88SSNGLEE343 pKa = 4.42DD344 pKa = 3.14QTPRR348 pKa = 11.84QNTQVANFTFIQRR361 pKa = 11.84RR362 pKa = 11.84SGDD365 pKa = 3.2QVIRR369 pKa = 11.84LRR371 pKa = 11.84GGTDD375 pKa = 2.93YY376 pKa = 11.6GLYY379 pKa = 9.99NGVVIDD385 pKa = 4.1QSSSGTPCIRR395 pKa = 11.84IDD397 pKa = 3.6DD398 pKa = 4.08QEE400 pKa = 4.46TIRR403 pKa = 11.84AANAGLDD410 pKa = 3.76DD411 pKa = 4.03VGPPRR416 pKa = 11.84FNSVALDD423 pKa = 3.52CQTNFRR429 pKa = 11.84AGSDD433 pKa = 3.58GVTVADD439 pKa = 3.91VEE441 pKa = 4.92AILTSGTGNTLDD453 pKa = 4.16YY454 pKa = 11.4DD455 pKa = 3.74NTIQNGYY462 pKa = 10.3LNGANEE468 pKa = 4.68DD469 pKa = 3.71GYY471 pKa = 11.12MPIFDD476 pKa = 4.02VTSLSSFFEE485 pKa = 4.18SASFIGAVSASNDD498 pKa = 2.81WTQGWTCDD506 pKa = 3.18SAAITFGNNTGNCLTLPVYY525 pKa = 10.6
Molecular weight: 54.38 kDa
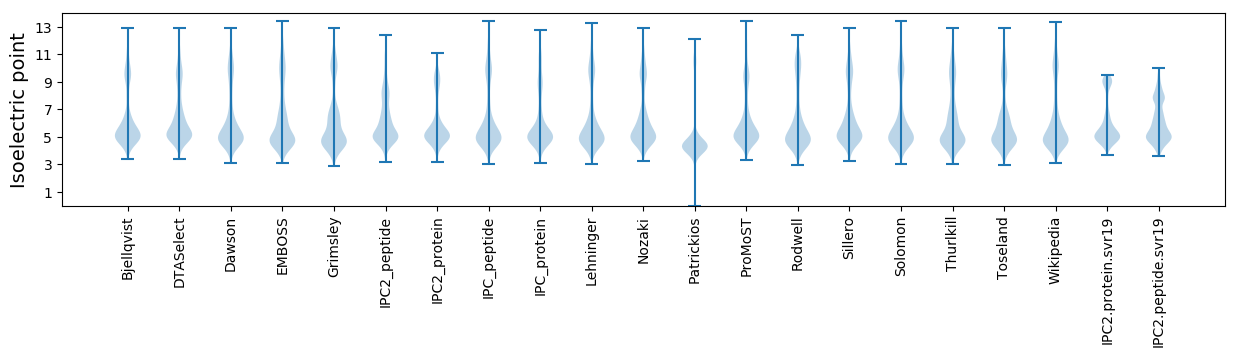
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N5CVL0|A0A3N5CVL0_9SPHN ATP synthase gamma chain OS=Aurantiacibacter spongiae OX=2488860 GN=atpG PE=3 SV=1
MM1 pKa = 7.22TFSHH5 pKa = 7.47PRR7 pKa = 11.84RR8 pKa = 11.84QPRR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84EE18 pKa = 3.49PRR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84TRR27 pKa = 11.84PRR29 pKa = 11.84ANPRR33 pKa = 11.84PPRR36 pKa = 11.84SATTVLPARR45 pKa = 11.84SPDD48 pKa = 3.21RR49 pKa = 11.84ARR51 pKa = 11.84PARR54 pKa = 11.84PAVRR58 pKa = 11.84RR59 pKa = 11.84HH60 pKa = 6.08RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TWPKK67 pKa = 9.07GRR69 pKa = 11.84TGRR72 pKa = 11.84RR73 pKa = 3.22
MM1 pKa = 7.22TFSHH5 pKa = 7.47PRR7 pKa = 11.84RR8 pKa = 11.84QPRR11 pKa = 11.84RR12 pKa = 11.84SRR14 pKa = 11.84RR15 pKa = 11.84SRR17 pKa = 11.84EE18 pKa = 3.49PRR20 pKa = 11.84PRR22 pKa = 11.84RR23 pKa = 11.84LRR25 pKa = 11.84TRR27 pKa = 11.84PRR29 pKa = 11.84ANPRR33 pKa = 11.84PPRR36 pKa = 11.84SATTVLPARR45 pKa = 11.84SPDD48 pKa = 3.21RR49 pKa = 11.84ARR51 pKa = 11.84PARR54 pKa = 11.84PAVRR58 pKa = 11.84RR59 pKa = 11.84HH60 pKa = 6.08RR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84TWPKK67 pKa = 9.07GRR69 pKa = 11.84TGRR72 pKa = 11.84RR73 pKa = 3.22
Molecular weight: 8.86 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
867789 |
41 |
1865 |
322.6 |
34.98 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.965 ± 0.067 | 0.836 ± 0.014 |
6.566 ± 0.037 | 6.144 ± 0.048 |
3.533 ± 0.028 | 8.963 ± 0.048 |
1.916 ± 0.022 | 4.743 ± 0.031 |
2.514 ± 0.039 | 9.758 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.475 ± 0.023 | 2.479 ± 0.025 |
5.202 ± 0.037 | 3.033 ± 0.029 |
7.841 ± 0.05 | 5.137 ± 0.036 |
5.191 ± 0.029 | 7.094 ± 0.035 |
1.43 ± 0.019 | 2.182 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
