
Mosquito circovirus
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.83
Get precalculated fractions of proteins
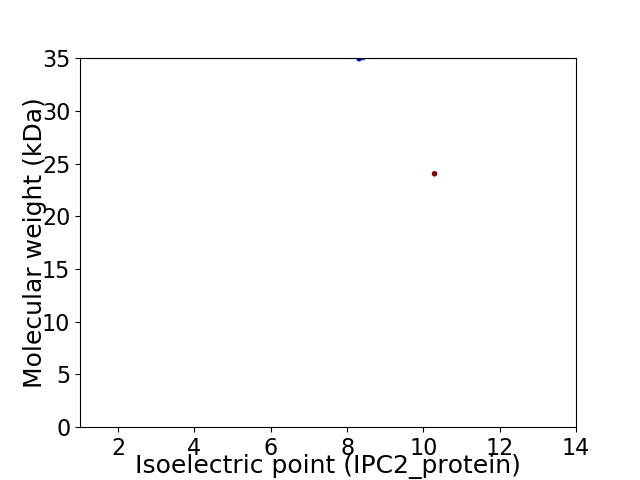
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5C3Z3|A0A0C5C3Z3_9CIRC ATP-dependent helicase Rep OS=Mosquito circovirus OX=1611039 PE=3 SV=1
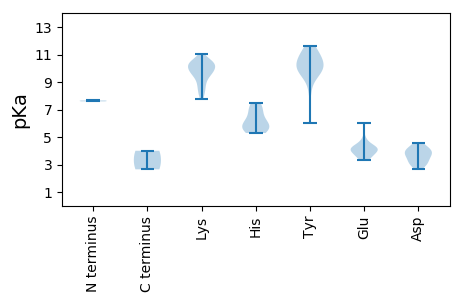
MM1 pKa = 7.59SLIQFVCWTLNNYY14 pKa = 9.81SEE16 pKa = 6.04DD17 pKa = 4.05EE18 pKa = 4.24YY19 pKa = 11.55QSLIEE24 pKa = 4.13FSEE27 pKa = 3.47WRR29 pKa = 11.84YY30 pKa = 9.9IVIGRR35 pKa = 11.84EE36 pKa = 3.93VGEE39 pKa = 4.37TGTPHH44 pKa = 5.54LQGYY48 pKa = 9.51GEE50 pKa = 4.06LKK52 pKa = 10.06KK53 pKa = 10.47RR54 pKa = 11.84KK55 pKa = 9.53KK56 pKa = 9.92FAALKK61 pKa = 9.41NLFPRR66 pKa = 11.84VHH68 pKa = 6.07WEE70 pKa = 3.43QRR72 pKa = 11.84RR73 pKa = 11.84ASRR76 pKa = 11.84DD77 pKa = 3.0AAANYY82 pKa = 8.24CKK84 pKa = 10.39KK85 pKa = 10.27EE86 pKa = 3.75GRR88 pKa = 11.84FEE90 pKa = 4.17EE91 pKa = 4.91RR92 pKa = 11.84GSLPKK97 pKa = 10.36NASEE101 pKa = 4.34KK102 pKa = 10.19SSKK105 pKa = 9.99EE106 pKa = 3.31AITRR110 pKa = 11.84VKK112 pKa = 10.33SGQTMRR118 pKa = 11.84AILDD122 pKa = 3.95DD123 pKa = 4.43PPNLSGIRR131 pKa = 11.84MCQIWLSYY139 pKa = 11.01NEE141 pKa = 4.09PKK143 pKa = 10.55RR144 pKa = 11.84NFKK147 pKa = 11.01SEE149 pKa = 3.97VFWYY153 pKa = 10.23YY154 pKa = 10.77GASGTGKK161 pKa = 8.35TKK163 pKa = 10.46LASEE167 pKa = 4.24QAGPDD172 pKa = 4.02AYY174 pKa = 9.78WHH176 pKa = 7.46DD177 pKa = 3.62GTKK180 pKa = 9.79WFDD183 pKa = 3.32GYY185 pKa = 10.9DD186 pKa = 3.35AHH188 pKa = 6.85EE189 pKa = 4.73HH190 pKa = 5.87VLLDD194 pKa = 4.21DD195 pKa = 3.82YY196 pKa = 11.56RR197 pKa = 11.84GGNMKK202 pKa = 10.39FNFLLKK208 pKa = 10.6FLDD211 pKa = 4.19RR212 pKa = 11.84YY213 pKa = 9.48PLRR216 pKa = 11.84LEE218 pKa = 4.06VKK220 pKa = 9.72GGYY223 pKa = 9.0RR224 pKa = 11.84QLLAKK229 pKa = 9.57KK230 pKa = 9.69IWVTSIKK237 pKa = 10.06HH238 pKa = 5.29PKK240 pKa = 9.13EE241 pKa = 3.73IYY243 pKa = 10.21SFSEE247 pKa = 3.53MDD249 pKa = 3.65EE250 pKa = 4.0PTEE253 pKa = 3.76QLLRR257 pKa = 11.84RR258 pKa = 11.84ITTIKK263 pKa = 10.33KK264 pKa = 8.32MCACSQCEE272 pKa = 3.58RR273 pKa = 11.84FYY275 pKa = 11.6GNRR278 pKa = 11.84FEE280 pKa = 4.31VSRR283 pKa = 11.84FAVCGNTSTNSDD295 pKa = 3.09TAANEE300 pKa = 4.09TT301 pKa = 4.0
MM1 pKa = 7.59SLIQFVCWTLNNYY14 pKa = 9.81SEE16 pKa = 6.04DD17 pKa = 4.05EE18 pKa = 4.24YY19 pKa = 11.55QSLIEE24 pKa = 4.13FSEE27 pKa = 3.47WRR29 pKa = 11.84YY30 pKa = 9.9IVIGRR35 pKa = 11.84EE36 pKa = 3.93VGEE39 pKa = 4.37TGTPHH44 pKa = 5.54LQGYY48 pKa = 9.51GEE50 pKa = 4.06LKK52 pKa = 10.06KK53 pKa = 10.47RR54 pKa = 11.84KK55 pKa = 9.53KK56 pKa = 9.92FAALKK61 pKa = 9.41NLFPRR66 pKa = 11.84VHH68 pKa = 6.07WEE70 pKa = 3.43QRR72 pKa = 11.84RR73 pKa = 11.84ASRR76 pKa = 11.84DD77 pKa = 3.0AAANYY82 pKa = 8.24CKK84 pKa = 10.39KK85 pKa = 10.27EE86 pKa = 3.75GRR88 pKa = 11.84FEE90 pKa = 4.17EE91 pKa = 4.91RR92 pKa = 11.84GSLPKK97 pKa = 10.36NASEE101 pKa = 4.34KK102 pKa = 10.19SSKK105 pKa = 9.99EE106 pKa = 3.31AITRR110 pKa = 11.84VKK112 pKa = 10.33SGQTMRR118 pKa = 11.84AILDD122 pKa = 3.95DD123 pKa = 4.43PPNLSGIRR131 pKa = 11.84MCQIWLSYY139 pKa = 11.01NEE141 pKa = 4.09PKK143 pKa = 10.55RR144 pKa = 11.84NFKK147 pKa = 11.01SEE149 pKa = 3.97VFWYY153 pKa = 10.23YY154 pKa = 10.77GASGTGKK161 pKa = 8.35TKK163 pKa = 10.46LASEE167 pKa = 4.24QAGPDD172 pKa = 4.02AYY174 pKa = 9.78WHH176 pKa = 7.46DD177 pKa = 3.62GTKK180 pKa = 9.79WFDD183 pKa = 3.32GYY185 pKa = 10.9DD186 pKa = 3.35AHH188 pKa = 6.85EE189 pKa = 4.73HH190 pKa = 5.87VLLDD194 pKa = 4.21DD195 pKa = 3.82YY196 pKa = 11.56RR197 pKa = 11.84GGNMKK202 pKa = 10.39FNFLLKK208 pKa = 10.6FLDD211 pKa = 4.19RR212 pKa = 11.84YY213 pKa = 9.48PLRR216 pKa = 11.84LEE218 pKa = 4.06VKK220 pKa = 9.72GGYY223 pKa = 9.0RR224 pKa = 11.84QLLAKK229 pKa = 9.57KK230 pKa = 9.69IWVTSIKK237 pKa = 10.06HH238 pKa = 5.29PKK240 pKa = 9.13EE241 pKa = 3.73IYY243 pKa = 10.21SFSEE247 pKa = 3.53MDD249 pKa = 3.65EE250 pKa = 4.0PTEE253 pKa = 3.76QLLRR257 pKa = 11.84RR258 pKa = 11.84ITTIKK263 pKa = 10.33KK264 pKa = 8.32MCACSQCEE272 pKa = 3.58RR273 pKa = 11.84FYY275 pKa = 11.6GNRR278 pKa = 11.84FEE280 pKa = 4.31VSRR283 pKa = 11.84FAVCGNTSTNSDD295 pKa = 3.09TAANEE300 pKa = 4.09TT301 pKa = 4.0
Molecular weight: 34.96 kDa
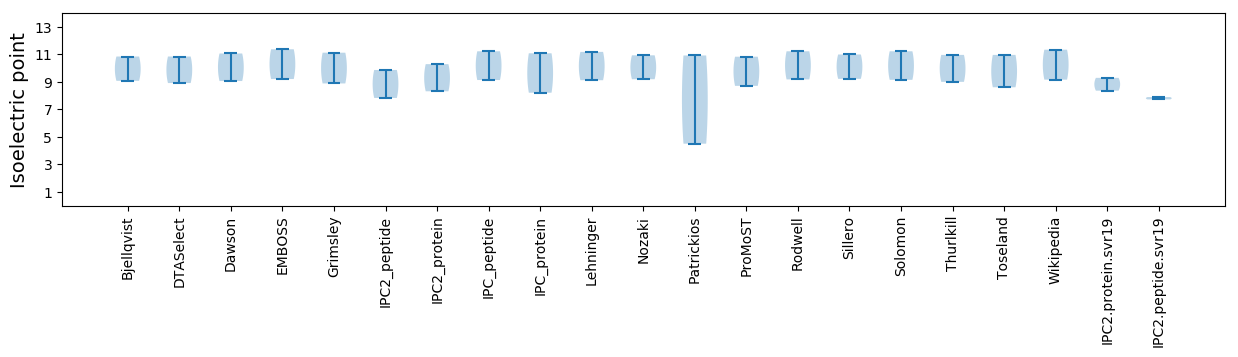
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5C3Z3|A0A0C5C3Z3_9CIRC ATP-dependent helicase Rep OS=Mosquito circovirus OX=1611039 PE=3 SV=1
MM1 pKa = 7.66AYY3 pKa = 9.97KK4 pKa = 10.63RR5 pKa = 11.84MRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84KK11 pKa = 7.8RR12 pKa = 11.84APRR15 pKa = 11.84KK16 pKa = 8.08RR17 pKa = 11.84SYY19 pKa = 10.28RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FNTSITRR33 pKa = 11.84TVLRR37 pKa = 11.84KK38 pKa = 9.77FFDD41 pKa = 3.68TSFSTTDD48 pKa = 2.7VGYY51 pKa = 11.0NYY53 pKa = 10.66LSDD56 pKa = 3.99NPKK59 pKa = 8.91GTPGWSYY66 pKa = 9.41YY67 pKa = 10.89ASSFHH72 pKa = 7.04HH73 pKa = 5.72YY74 pKa = 9.79RR75 pKa = 11.84VVGLALKK82 pKa = 8.63FVPKK86 pKa = 9.77ATTAPMEE93 pKa = 4.4TGMVHH98 pKa = 6.26YY99 pKa = 10.46DD100 pKa = 2.93SSLFFARR107 pKa = 11.84DD108 pKa = 3.19ISPVVWAEE116 pKa = 3.92TGNNLINKK124 pKa = 8.7ALSIPNMKK132 pKa = 9.85QRR134 pKa = 11.84TLTRR138 pKa = 11.84SFSLYY143 pKa = 10.02FPMRR147 pKa = 11.84KK148 pKa = 9.34SSPTTYY154 pKa = 10.1QATAGGYY161 pKa = 10.52LSTDD165 pKa = 3.62DD166 pKa = 4.6PKK168 pKa = 9.11TTQLVHH174 pKa = 5.53MVSEE178 pKa = 4.45KK179 pKa = 10.22MSFAIPGRR187 pKa = 11.84FYY189 pKa = 11.05LSLYY193 pKa = 10.23IRR195 pKa = 11.84FKK197 pKa = 10.3NKK199 pKa = 9.75RR200 pKa = 11.84YY201 pKa = 6.03TTTPVV206 pKa = 2.65
MM1 pKa = 7.66AYY3 pKa = 9.97KK4 pKa = 10.63RR5 pKa = 11.84MRR7 pKa = 11.84RR8 pKa = 11.84SRR10 pKa = 11.84KK11 pKa = 7.8RR12 pKa = 11.84APRR15 pKa = 11.84KK16 pKa = 8.08RR17 pKa = 11.84SYY19 pKa = 10.28RR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FNTSITRR33 pKa = 11.84TVLRR37 pKa = 11.84KK38 pKa = 9.77FFDD41 pKa = 3.68TSFSTTDD48 pKa = 2.7VGYY51 pKa = 11.0NYY53 pKa = 10.66LSDD56 pKa = 3.99NPKK59 pKa = 8.91GTPGWSYY66 pKa = 9.41YY67 pKa = 10.89ASSFHH72 pKa = 7.04HH73 pKa = 5.72YY74 pKa = 9.79RR75 pKa = 11.84VVGLALKK82 pKa = 8.63FVPKK86 pKa = 9.77ATTAPMEE93 pKa = 4.4TGMVHH98 pKa = 6.26YY99 pKa = 10.46DD100 pKa = 2.93SSLFFARR107 pKa = 11.84DD108 pKa = 3.19ISPVVWAEE116 pKa = 3.92TGNNLINKK124 pKa = 8.7ALSIPNMKK132 pKa = 9.85QRR134 pKa = 11.84TLTRR138 pKa = 11.84SFSLYY143 pKa = 10.02FPMRR147 pKa = 11.84KK148 pKa = 9.34SSPTTYY154 pKa = 10.1QATAGGYY161 pKa = 10.52LSTDD165 pKa = 3.62DD166 pKa = 4.6PKK168 pKa = 9.11TTQLVHH174 pKa = 5.53MVSEE178 pKa = 4.45KK179 pKa = 10.22MSFAIPGRR187 pKa = 11.84FYY189 pKa = 11.05LSLYY193 pKa = 10.23IRR195 pKa = 11.84FKK197 pKa = 10.3NKK199 pKa = 9.75RR200 pKa = 11.84YY201 pKa = 6.03TTTPVV206 pKa = 2.65
Molecular weight: 24.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
507 |
206 |
301 |
253.5 |
29.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.114 ± 0.177 | 1.381 ± 0.846 |
3.945 ± 0.335 | 5.523 ± 2.49 |
5.72 ± 0.659 | 5.917 ± 0.948 |
1.972 ± 0.019 | 3.748 ± 0.511 |
7.692 ± 0.549 | 7.101 ± 0.484 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.761 ± 0.687 | 4.339 ± 0.279 |
4.339 ± 0.91 | 2.564 ± 0.678 |
8.876 ± 1.402 | 8.481 ± 1.049 |
7.495 ± 1.95 | 4.339 ± 0.613 |
1.972 ± 0.613 | 5.72 ± 0.659 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
