
Aspergillus heteromorphus CBS 117.55
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Aspergillus; Aspergillus heteromorphus
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
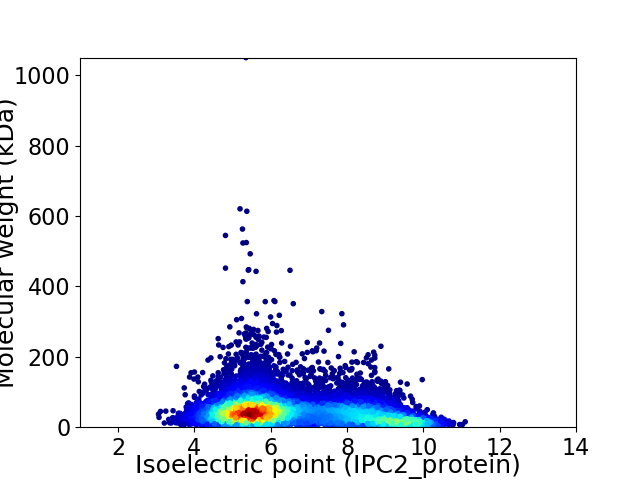
Virtual 2D-PAGE plot for 10837 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A317V505|A0A317V505_9EURO Isochorismatase hydrolase OS=Aspergillus heteromorphus CBS 117.55 OX=1448321 GN=BO70DRAFT_366221 PE=3 SV=1
DDD2 pKa = 4.18DD3 pKa = 4.43GDDD6 pKa = 3.78DD7 pKa = 4.0GDDD10 pKa = 3.55DD11 pKa = 3.93GDDD14 pKa = 3.55DD15 pKa = 3.93GDDD18 pKa = 3.55DD19 pKa = 3.93GDDD22 pKa = 3.55DD23 pKa = 3.93GDDD26 pKa = 3.55DD27 pKa = 3.93GDDD30 pKa = 3.55DD31 pKa = 3.93GDDD34 pKa = 3.55DD35 pKa = 3.93GDDD38 pKa = 3.55DD39 pKa = 3.93GDDD42 pKa = 3.58DD43 pKa = 3.94GDDD46 pKa = 3.71DD47 pKa = 3.99DDD49 pKa = 5.25DDD51 pKa = 4.06GDDD54 pKa = 3.61YY55 pKa = 9.91DDD57 pKa = 4.85DDD59 pKa = 4.02GDDD62 pKa = 4.0DD63 pKa = 3.67VASSTYYY70 pKa = 8.64TPATSSSAAVSDDD83 pKa = 5.65DD84 pKa = 3.87EE85 pKa = 5.13CSPQCTEEE93 pKa = 5.21ISNDDD98 pKa = 3.34TDDD101 pKa = 3.66SSASSKKK108 pKa = 9.61SNWFDDD114 pKa = 3.21GLSKKK119 pKa = 10.21TSDDD123 pKa = 3.51YY124 pKa = 9.44TWMLTQMRR132 pKa = 11.84TGPLLTKKK140 pKa = 9.97DD141 pKa = 3.47SGMTTSEEE149 pKa = 3.85VVFDDD154 pKa = 3.48SAINFSQTNIRR165 pKa = 11.84GCTVVIVVSKKK176 pKa = 9.06YY177 pKa = 10.34AYYY180 pKa = 9.52AHHH183 pKa = 6.57WEEE186 pKa = 5.06DD187 pKa = 3.53GFVSQTNDDD196 pKa = 2.38TTAVLNEEE204 pKa = 4.31EEE206 pKa = 4.28QNTVLTYYY214 pKa = 8.94TNDDD218 pKa = 3.64NLKKK222 pKa = 9.16PQLTGSTAYYY232 pKa = 9.25DDD234 pKa = 4.44GTQVYYY240 pKa = 10.22GTPQAQSGKKK250 pKa = 8.33TAKKK254 pKa = 10.75YY255 pKa = 7.66AQVSRR260 pKa = 11.84VGITVNEEE268 pKa = 4.0LGLSAAPTEEE278 pKa = 4.66YY279 pKa = 10.7YY280 pKa = 10.9YY281 pKa = 10.27TEEE284 pKa = 3.89EE285 pKa = 3.81AGEEE289 pKa = 4.36KKK291 pKa = 9.32LFQFPNSCDDD301 pKa = 3.43KK302 pKa = 10.77LWQIWYYY309 pKa = 10.23DDD311 pKa = 4.22DD312 pKa = 3.7NSALWASSQNGACSSAPAPASASSSVTAVRR342 pKa = 11.84TPPFPIFTDDD352 pKa = 3.49STLIARR358 pKa = 11.84PRR360 pKa = 11.84TTTQAYYY367 pKa = 8.17
DDD2 pKa = 4.18DD3 pKa = 4.43GDDD6 pKa = 3.78DD7 pKa = 4.0GDDD10 pKa = 3.55DD11 pKa = 3.93GDDD14 pKa = 3.55DD15 pKa = 3.93GDDD18 pKa = 3.55DD19 pKa = 3.93GDDD22 pKa = 3.55DD23 pKa = 3.93GDDD26 pKa = 3.55DD27 pKa = 3.93GDDD30 pKa = 3.55DD31 pKa = 3.93GDDD34 pKa = 3.55DD35 pKa = 3.93GDDD38 pKa = 3.55DD39 pKa = 3.93GDDD42 pKa = 3.58DD43 pKa = 3.94GDDD46 pKa = 3.71DD47 pKa = 3.99DDD49 pKa = 5.25DDD51 pKa = 4.06GDDD54 pKa = 3.61YY55 pKa = 9.91DDD57 pKa = 4.85DDD59 pKa = 4.02GDDD62 pKa = 4.0DD63 pKa = 3.67VASSTYYY70 pKa = 8.64TPATSSSAAVSDDD83 pKa = 5.65DD84 pKa = 3.87EE85 pKa = 5.13CSPQCTEEE93 pKa = 5.21ISNDDD98 pKa = 3.34TDDD101 pKa = 3.66SSASSKKK108 pKa = 9.61SNWFDDD114 pKa = 3.21GLSKKK119 pKa = 10.21TSDDD123 pKa = 3.51YY124 pKa = 9.44TWMLTQMRR132 pKa = 11.84TGPLLTKKK140 pKa = 9.97DD141 pKa = 3.47SGMTTSEEE149 pKa = 3.85VVFDDD154 pKa = 3.48SAINFSQTNIRR165 pKa = 11.84GCTVVIVVSKKK176 pKa = 9.06YY177 pKa = 10.34AYYY180 pKa = 9.52AHHH183 pKa = 6.57WEEE186 pKa = 5.06DD187 pKa = 3.53GFVSQTNDDD196 pKa = 2.38TTAVLNEEE204 pKa = 4.31EEE206 pKa = 4.28QNTVLTYYY214 pKa = 8.94TNDDD218 pKa = 3.64NLKKK222 pKa = 9.16PQLTGSTAYYY232 pKa = 9.25DDD234 pKa = 4.44GTQVYYY240 pKa = 10.22GTPQAQSGKKK250 pKa = 8.33TAKKK254 pKa = 10.75YY255 pKa = 7.66AQVSRR260 pKa = 11.84VGITVNEEE268 pKa = 4.0LGLSAAPTEEE278 pKa = 4.66YY279 pKa = 10.7YY280 pKa = 10.9YY281 pKa = 10.27TEEE284 pKa = 3.89EE285 pKa = 3.81AGEEE289 pKa = 4.36KKK291 pKa = 9.32LFQFPNSCDDD301 pKa = 3.43KK302 pKa = 10.77LWQIWYYY309 pKa = 10.23DDD311 pKa = 4.22DD312 pKa = 3.7NSALWASSQNGACSSAPAPASASSSVTAVRR342 pKa = 11.84TPPFPIFTDDD352 pKa = 3.49STLIARR358 pKa = 11.84PRR360 pKa = 11.84TTTQAYYY367 pKa = 8.17
Molecular weight: 39.02 kDa
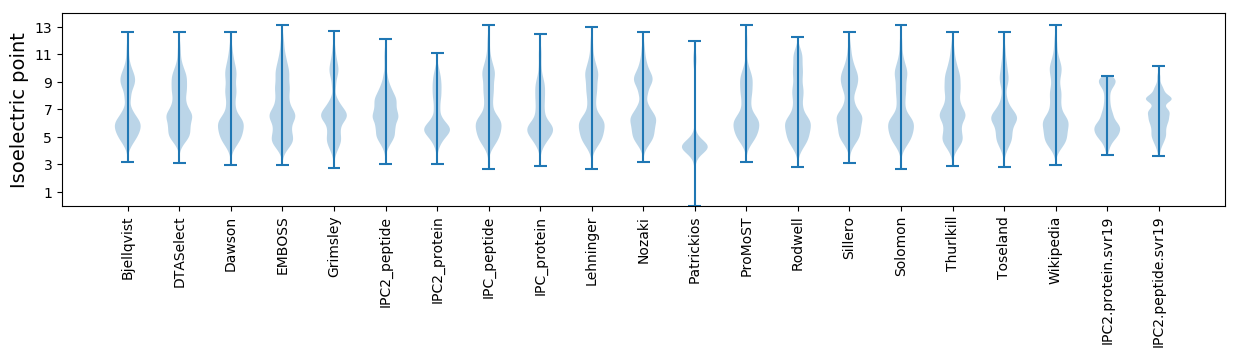
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A317WLR7|A0A317WLR7_9EURO Uncharacterized protein OS=Aspergillus heteromorphus CBS 117.55 OX=1448321 GN=BO70DRAFT_193952 PE=4 SV=1
MM1 pKa = 7.09FCFRR5 pKa = 11.84SRR7 pKa = 11.84ALPAAIRR14 pKa = 11.84AYY16 pKa = 10.0AAPTARR22 pKa = 11.84LSSAPITPIATSLSTPLRR40 pKa = 11.84TLSSTTLSSLRR51 pKa = 11.84QHH53 pKa = 6.67IPSQTLSSSSTTFSSLRR70 pKa = 11.84PLLTTSPFTAQQTRR84 pKa = 11.84SFSASASLGGKK95 pKa = 9.56RR96 pKa = 11.84NTYY99 pKa = 9.03NPSRR103 pKa = 11.84RR104 pKa = 11.84VQKK107 pKa = 10.1RR108 pKa = 11.84RR109 pKa = 11.84HH110 pKa = 5.44GFLSRR115 pKa = 11.84LRR117 pKa = 11.84TRR119 pKa = 11.84GGRR122 pKa = 11.84NILMRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84QKK131 pKa = 10.54GRR133 pKa = 11.84KK134 pKa = 7.54NLSWW138 pKa = 4.16
MM1 pKa = 7.09FCFRR5 pKa = 11.84SRR7 pKa = 11.84ALPAAIRR14 pKa = 11.84AYY16 pKa = 10.0AAPTARR22 pKa = 11.84LSSAPITPIATSLSTPLRR40 pKa = 11.84TLSSTTLSSLRR51 pKa = 11.84QHH53 pKa = 6.67IPSQTLSSSSTTFSSLRR70 pKa = 11.84PLLTTSPFTAQQTRR84 pKa = 11.84SFSASASLGGKK95 pKa = 9.56RR96 pKa = 11.84NTYY99 pKa = 9.03NPSRR103 pKa = 11.84RR104 pKa = 11.84VQKK107 pKa = 10.1RR108 pKa = 11.84RR109 pKa = 11.84HH110 pKa = 5.44GFLSRR115 pKa = 11.84LRR117 pKa = 11.84TRR119 pKa = 11.84GGRR122 pKa = 11.84NILMRR127 pKa = 11.84RR128 pKa = 11.84RR129 pKa = 11.84QKK131 pKa = 10.54GRR133 pKa = 11.84KK134 pKa = 7.54NLSWW138 pKa = 4.16
Molecular weight: 15.32 kDa
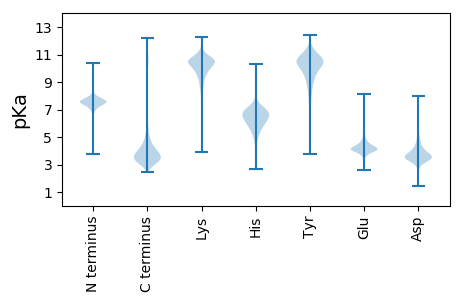
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4977815 |
49 |
9562 |
459.3 |
50.78 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.592 ± 0.023 | 1.281 ± 0.009 |
5.625 ± 0.016 | 6.009 ± 0.026 |
3.718 ± 0.013 | 6.915 ± 0.023 |
2.507 ± 0.011 | 4.785 ± 0.016 |
4.252 ± 0.022 | 9.142 ± 0.026 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.162 ± 0.008 | 3.419 ± 0.013 |
6.333 ± 0.028 | 3.971 ± 0.016 |
6.3 ± 0.021 | 8.387 ± 0.028 |
6.064 ± 0.013 | 6.252 ± 0.016 |
1.474 ± 0.009 | 2.814 ± 0.012 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
