
Lake Sarah-associated circular virus-39
Taxonomy: Viruses; unclassified viruses
Average proteome isoelectric point is 7.36
Get precalculated fractions of proteins
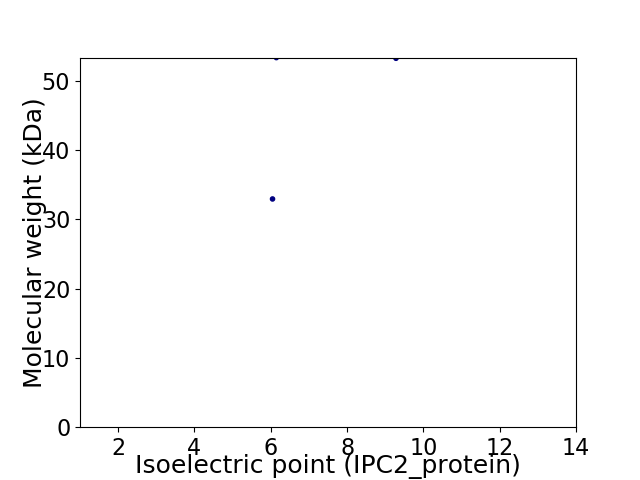
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
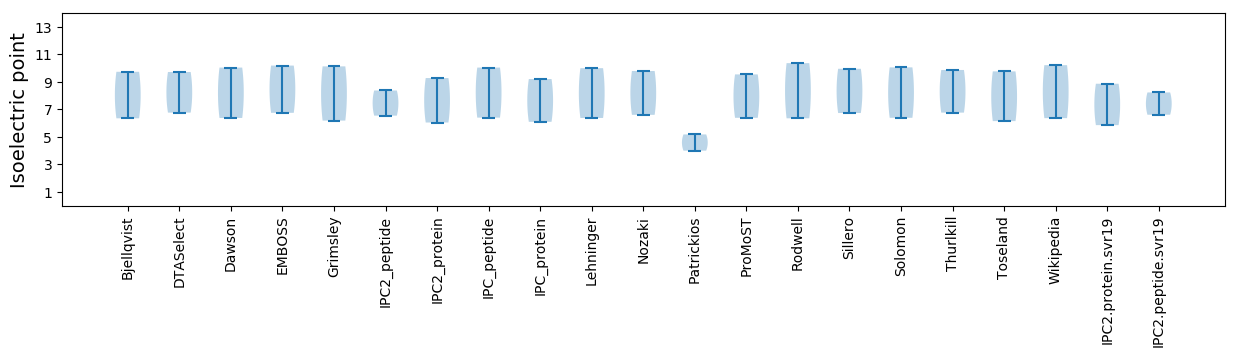
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140AQQ0|A0A140AQQ0_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-39 OX=1685767 PE=3 SV=1
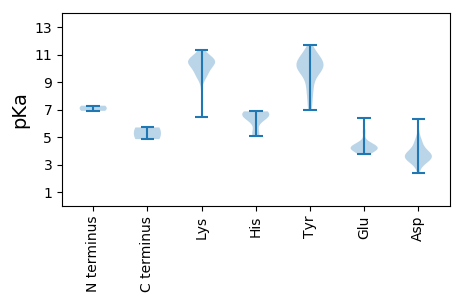
MM1 pKa = 7.28YY2 pKa = 8.05MARR5 pKa = 11.84NRR7 pKa = 11.84NFVFTWNNYY16 pKa = 8.48TEE18 pKa = 5.43DD19 pKa = 3.44SEE21 pKa = 4.53QFLRR25 pKa = 11.84SRR27 pKa = 11.84VDD29 pKa = 3.14EE30 pKa = 4.27GVIKK34 pKa = 10.77YY35 pKa = 10.09VGYY38 pKa = 10.18CKK40 pKa = 10.31EE41 pKa = 4.04IAPSTGTPHH50 pKa = 6.75LQGFCSFEE58 pKa = 4.11NKK60 pKa = 10.01KK61 pKa = 10.37SVQQVRR67 pKa = 11.84VILVGCHH74 pKa = 5.07VEE76 pKa = 4.22TMLGSISQNEE86 pKa = 4.54DD87 pKa = 3.37YY88 pKa = 10.7CSKK91 pKa = 10.83AGEE94 pKa = 4.4LINYY98 pKa = 7.43GVQPVTNDD106 pKa = 2.79NKK108 pKa = 10.43GRR110 pKa = 11.84AEE112 pKa = 3.73QLRR115 pKa = 11.84WQRR118 pKa = 11.84ARR120 pKa = 11.84EE121 pKa = 4.03SAKK124 pKa = 10.49LGQFDD129 pKa = 6.3DD130 pKa = 3.52IDD132 pKa = 5.38ADD134 pKa = 3.61IFIRR138 pKa = 11.84CYY140 pKa = 9.79STLKK144 pKa = 10.54RR145 pKa = 11.84IRR147 pKa = 11.84TDD149 pKa = 3.33YY150 pKa = 8.97TAKK153 pKa = 10.5PPPEE157 pKa = 5.34DD158 pKa = 3.51VTCYY162 pKa = 9.98WIYY165 pKa = 11.16GPTGTGKK172 pKa = 9.3SHH174 pKa = 6.42SVEE177 pKa = 4.14TTFPLCYY184 pKa = 9.29KK185 pKa = 10.79KK186 pKa = 10.9NMDD189 pKa = 3.76DD190 pKa = 4.02PKK192 pKa = 10.73WFDD195 pKa = 3.36GYY197 pKa = 10.93QGEE200 pKa = 4.4EE201 pKa = 3.77TVYY204 pKa = 11.22LEE206 pKa = 6.39DD207 pKa = 3.45IDD209 pKa = 4.66KK210 pKa = 11.25YY211 pKa = 7.36QVKK214 pKa = 9.65WGGLLKK220 pKa = 10.55RR221 pKa = 11.84LADD224 pKa = 3.96RR225 pKa = 11.84WPLLVNTKK233 pKa = 10.51GSMQYY238 pKa = 10.04IRR240 pKa = 11.84PKK242 pKa = 10.43RR243 pKa = 11.84IIVTSNYY250 pKa = 10.12NLDD253 pKa = 4.74EE254 pKa = 4.28IWSDD258 pKa = 3.88SGTLDD263 pKa = 3.38PLLRR267 pKa = 11.84RR268 pKa = 11.84FTVILKK274 pKa = 9.63EE275 pKa = 4.15SQEE278 pKa = 4.0QVIDD282 pKa = 3.71FTT284 pKa = 4.84
MM1 pKa = 7.28YY2 pKa = 8.05MARR5 pKa = 11.84NRR7 pKa = 11.84NFVFTWNNYY16 pKa = 8.48TEE18 pKa = 5.43DD19 pKa = 3.44SEE21 pKa = 4.53QFLRR25 pKa = 11.84SRR27 pKa = 11.84VDD29 pKa = 3.14EE30 pKa = 4.27GVIKK34 pKa = 10.77YY35 pKa = 10.09VGYY38 pKa = 10.18CKK40 pKa = 10.31EE41 pKa = 4.04IAPSTGTPHH50 pKa = 6.75LQGFCSFEE58 pKa = 4.11NKK60 pKa = 10.01KK61 pKa = 10.37SVQQVRR67 pKa = 11.84VILVGCHH74 pKa = 5.07VEE76 pKa = 4.22TMLGSISQNEE86 pKa = 4.54DD87 pKa = 3.37YY88 pKa = 10.7CSKK91 pKa = 10.83AGEE94 pKa = 4.4LINYY98 pKa = 7.43GVQPVTNDD106 pKa = 2.79NKK108 pKa = 10.43GRR110 pKa = 11.84AEE112 pKa = 3.73QLRR115 pKa = 11.84WQRR118 pKa = 11.84ARR120 pKa = 11.84EE121 pKa = 4.03SAKK124 pKa = 10.49LGQFDD129 pKa = 6.3DD130 pKa = 3.52IDD132 pKa = 5.38ADD134 pKa = 3.61IFIRR138 pKa = 11.84CYY140 pKa = 9.79STLKK144 pKa = 10.54RR145 pKa = 11.84IRR147 pKa = 11.84TDD149 pKa = 3.33YY150 pKa = 8.97TAKK153 pKa = 10.5PPPEE157 pKa = 5.34DD158 pKa = 3.51VTCYY162 pKa = 9.98WIYY165 pKa = 11.16GPTGTGKK172 pKa = 9.3SHH174 pKa = 6.42SVEE177 pKa = 4.14TTFPLCYY184 pKa = 9.29KK185 pKa = 10.79KK186 pKa = 10.9NMDD189 pKa = 3.76DD190 pKa = 4.02PKK192 pKa = 10.73WFDD195 pKa = 3.36GYY197 pKa = 10.93QGEE200 pKa = 4.4EE201 pKa = 3.77TVYY204 pKa = 11.22LEE206 pKa = 6.39DD207 pKa = 3.45IDD209 pKa = 4.66KK210 pKa = 11.25YY211 pKa = 7.36QVKK214 pKa = 9.65WGGLLKK220 pKa = 10.55RR221 pKa = 11.84LADD224 pKa = 3.96RR225 pKa = 11.84WPLLVNTKK233 pKa = 10.51GSMQYY238 pKa = 10.04IRR240 pKa = 11.84PKK242 pKa = 10.43RR243 pKa = 11.84IIVTSNYY250 pKa = 10.12NLDD253 pKa = 4.74EE254 pKa = 4.28IWSDD258 pKa = 3.88SGTLDD263 pKa = 3.38PLLRR267 pKa = 11.84RR268 pKa = 11.84FTVILKK274 pKa = 9.63EE275 pKa = 4.15SQEE278 pKa = 4.0QVIDD282 pKa = 3.71FTT284 pKa = 4.84
Molecular weight: 32.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140AQQ0|A0A140AQQ0_9VIRU ATP-dependent helicase Rep OS=Lake Sarah-associated circular virus-39 OX=1685767 PE=3 SV=1
MM1 pKa = 6.9KK2 pKa = 10.2RR3 pKa = 11.84KK4 pKa = 9.37YY5 pKa = 9.95RR6 pKa = 11.84SSDD9 pKa = 3.07SYY11 pKa = 11.69LPSTLTNSSPYY22 pKa = 9.73VRR24 pKa = 11.84RR25 pKa = 11.84TKK27 pKa = 10.09RR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 6.49TRR32 pKa = 11.84RR33 pKa = 11.84VSIDD37 pKa = 3.02YY38 pKa = 10.27GRR40 pKa = 11.84RR41 pKa = 11.84SSTSSGGLTNYY52 pKa = 8.28TSSSGSSVGRR62 pKa = 11.84AVASASGAALGFIHH76 pKa = 6.57MNVPGAVASYY86 pKa = 10.25KK87 pKa = 10.42KK88 pKa = 10.49AGEE91 pKa = 4.24FYY93 pKa = 10.73DD94 pKa = 3.75YY95 pKa = 10.4MNSKK99 pKa = 8.38TVSQTKK105 pKa = 10.03SSSNNMYY112 pKa = 9.93SGKK115 pKa = 9.59FRR117 pKa = 11.84KK118 pKa = 9.27PKK120 pKa = 9.85PFKK123 pKa = 10.15QSAMMKK129 pKa = 9.69AQTDD133 pKa = 4.83GYY135 pKa = 10.96LSTQEE140 pKa = 3.92QYY142 pKa = 10.32GTVTDD147 pKa = 4.23PNGVYY152 pKa = 10.32IFQSTFNSAQMALAITTALVRR173 pKa = 11.84KK174 pKa = 9.16LFKK177 pKa = 10.56KK178 pKa = 10.65AGITVSDD185 pKa = 3.95RR186 pKa = 11.84DD187 pKa = 3.33GSLAFFGVYY196 pKa = 9.96NADD199 pKa = 3.57GFKK202 pKa = 11.12LEE204 pKa = 4.13MVEE207 pKa = 4.12YY208 pKa = 11.02NPITGANSPAITYY221 pKa = 6.98TTVAADD227 pKa = 4.3TITTVVANSTFVTTFTNYY245 pKa = 9.92LNKK248 pKa = 9.58ATEE251 pKa = 4.26HH252 pKa = 6.87PPVSMTLYY260 pKa = 10.5SSDD263 pKa = 4.42RR264 pKa = 11.84NGLDD268 pKa = 3.59SNWRR272 pKa = 11.84LCSNLDD278 pKa = 3.58LNAEE282 pKa = 4.21YY283 pKa = 9.15LTVVANSTMVLQNQTKK299 pKa = 10.28AVISASNDD307 pKa = 2.37IDD309 pKa = 4.7RR310 pKa = 11.84NDD312 pKa = 3.57VQPLNVTMYY321 pKa = 10.71KK322 pKa = 10.51FKK324 pKa = 10.99NDD326 pKa = 3.79PKK328 pKa = 10.86LKK330 pKa = 10.61AIGNRR335 pKa = 11.84PSGSSKK341 pKa = 9.87TEE343 pKa = 4.51LIQWQGVGYY352 pKa = 10.29SGTRR356 pKa = 11.84LLRR359 pKa = 11.84AQDD362 pKa = 3.68FVNTSFQNPPPPGIFSNLTGKK383 pKa = 9.31SHH385 pKa = 6.49STLQPGQMKK394 pKa = 10.67KK395 pKa = 10.47NFLTHH400 pKa = 5.34TFKK403 pKa = 11.33GKK405 pKa = 10.35LSNILPKK412 pKa = 9.9MRR414 pKa = 11.84MEE416 pKa = 4.18QFGLSPIKK424 pKa = 10.53IHH426 pKa = 6.86GVYY429 pKa = 10.69CNTNLFCFQEE439 pKa = 4.39RR440 pKa = 11.84MRR442 pKa = 11.84TSTTNPIIVAYY453 pKa = 8.52EE454 pKa = 3.89HH455 pKa = 6.75EE456 pKa = 4.35LDD458 pKa = 3.95IGVFCATYY466 pKa = 9.53HH467 pKa = 6.42PKK469 pKa = 10.07IAMQASLDD477 pKa = 3.97SMDD480 pKa = 3.2VSLL483 pKa = 5.7
MM1 pKa = 6.9KK2 pKa = 10.2RR3 pKa = 11.84KK4 pKa = 9.37YY5 pKa = 9.95RR6 pKa = 11.84SSDD9 pKa = 3.07SYY11 pKa = 11.69LPSTLTNSSPYY22 pKa = 9.73VRR24 pKa = 11.84RR25 pKa = 11.84TKK27 pKa = 10.09RR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 6.49TRR32 pKa = 11.84RR33 pKa = 11.84VSIDD37 pKa = 3.02YY38 pKa = 10.27GRR40 pKa = 11.84RR41 pKa = 11.84SSTSSGGLTNYY52 pKa = 8.28TSSSGSSVGRR62 pKa = 11.84AVASASGAALGFIHH76 pKa = 6.57MNVPGAVASYY86 pKa = 10.25KK87 pKa = 10.42KK88 pKa = 10.49AGEE91 pKa = 4.24FYY93 pKa = 10.73DD94 pKa = 3.75YY95 pKa = 10.4MNSKK99 pKa = 8.38TVSQTKK105 pKa = 10.03SSSNNMYY112 pKa = 9.93SGKK115 pKa = 9.59FRR117 pKa = 11.84KK118 pKa = 9.27PKK120 pKa = 9.85PFKK123 pKa = 10.15QSAMMKK129 pKa = 9.69AQTDD133 pKa = 4.83GYY135 pKa = 10.96LSTQEE140 pKa = 3.92QYY142 pKa = 10.32GTVTDD147 pKa = 4.23PNGVYY152 pKa = 10.32IFQSTFNSAQMALAITTALVRR173 pKa = 11.84KK174 pKa = 9.16LFKK177 pKa = 10.56KK178 pKa = 10.65AGITVSDD185 pKa = 3.95RR186 pKa = 11.84DD187 pKa = 3.33GSLAFFGVYY196 pKa = 9.96NADD199 pKa = 3.57GFKK202 pKa = 11.12LEE204 pKa = 4.13MVEE207 pKa = 4.12YY208 pKa = 11.02NPITGANSPAITYY221 pKa = 6.98TTVAADD227 pKa = 4.3TITTVVANSTFVTTFTNYY245 pKa = 9.92LNKK248 pKa = 9.58ATEE251 pKa = 4.26HH252 pKa = 6.87PPVSMTLYY260 pKa = 10.5SSDD263 pKa = 4.42RR264 pKa = 11.84NGLDD268 pKa = 3.59SNWRR272 pKa = 11.84LCSNLDD278 pKa = 3.58LNAEE282 pKa = 4.21YY283 pKa = 9.15LTVVANSTMVLQNQTKK299 pKa = 10.28AVISASNDD307 pKa = 2.37IDD309 pKa = 4.7RR310 pKa = 11.84NDD312 pKa = 3.57VQPLNVTMYY321 pKa = 10.71KK322 pKa = 10.51FKK324 pKa = 10.99NDD326 pKa = 3.79PKK328 pKa = 10.86LKK330 pKa = 10.61AIGNRR335 pKa = 11.84PSGSSKK341 pKa = 9.87TEE343 pKa = 4.51LIQWQGVGYY352 pKa = 10.29SGTRR356 pKa = 11.84LLRR359 pKa = 11.84AQDD362 pKa = 3.68FVNTSFQNPPPPGIFSNLTGKK383 pKa = 9.31SHH385 pKa = 6.49STLQPGQMKK394 pKa = 10.67KK395 pKa = 10.47NFLTHH400 pKa = 5.34TFKK403 pKa = 11.33GKK405 pKa = 10.35LSNILPKK412 pKa = 9.9MRR414 pKa = 11.84MEE416 pKa = 4.18QFGLSPIKK424 pKa = 10.53IHH426 pKa = 6.86GVYY429 pKa = 10.69CNTNLFCFQEE439 pKa = 4.39RR440 pKa = 11.84MRR442 pKa = 11.84TSTTNPIIVAYY453 pKa = 8.52EE454 pKa = 3.89HH455 pKa = 6.75EE456 pKa = 4.35LDD458 pKa = 3.95IGVFCATYY466 pKa = 9.53HH467 pKa = 6.42PKK469 pKa = 10.07IAMQASLDD477 pKa = 3.97SMDD480 pKa = 3.2VSLL483 pKa = 5.7
Molecular weight: 53.26 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
767 |
284 |
483 |
383.5 |
43.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.476 ± 1.4 | 1.434 ± 0.625 |
5.215 ± 1.109 | 3.781 ± 1.552 |
4.302 ± 0.26 | 6.519 ± 0.104 |
1.304 ± 0.15 | 4.824 ± 0.705 |
6.649 ± 0.025 | 6.91 ± 0.08 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.868 ± 0.672 | 5.997 ± 0.862 |
4.433 ± 0.126 | 4.302 ± 0.381 |
5.346 ± 0.602 | 9.387 ± 2.064 |
8.605 ± 0.948 | 6.389 ± 0.183 |
1.173 ± 0.784 | 5.085 ± 0.333 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
