
Hoyosella subflava (strain DSM 45089 / JCM 17490 / NBRC 109087 / DQS3-9A1) (Amycolicicoccus subflavus)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Corynebacteriales; Mycobacteriaceae; Hoyosella; Hoyosella subflava
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
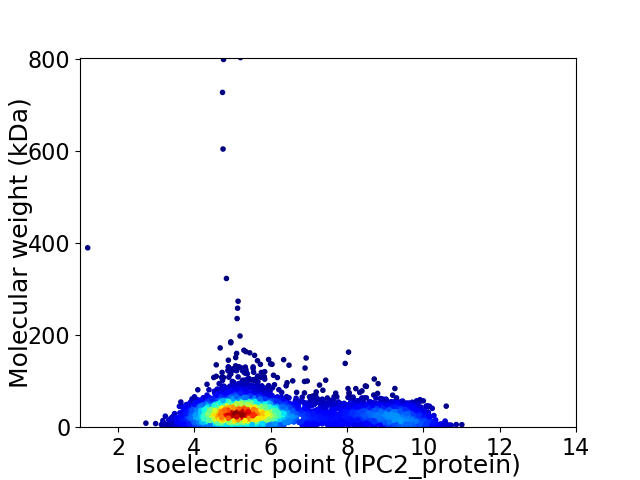
Virtual 2D-PAGE plot for 4703 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6EHV3|F6EHV3_HOYSD TetR-family transcriptional regulator OS=Hoyosella subflava (strain DSM 45089 / JCM 17490 / NBRC 109087 / DQS3-9A1) OX=443218 GN=AS9A_0444 PE=4 SV=1
MM1 pKa = 8.14DD2 pKa = 5.57DD3 pKa = 4.44EE4 pKa = 5.0EE5 pKa = 5.96DD6 pKa = 3.46VTAGVGVTEE15 pKa = 4.92EE16 pKa = 4.3PCPDD20 pKa = 4.05AVNADD25 pKa = 3.62NGCIYY30 pKa = 10.74LGVLSDD36 pKa = 4.45LEE38 pKa = 4.39GGPFAEE44 pKa = 5.77LGAQIQQGQLAFWRR58 pKa = 11.84SVNEE62 pKa = 4.11DD63 pKa = 3.29GGIAGYY69 pKa = 10.49DD70 pKa = 3.46VDD72 pKa = 3.15ISRR75 pKa = 11.84YY76 pKa = 6.69TRR78 pKa = 11.84NTAYY82 pKa = 10.63SVDD85 pKa = 3.55QHH87 pKa = 5.26ATEE90 pKa = 4.15YY91 pKa = 10.96QDD93 pKa = 4.28IEE95 pKa = 4.31PHH97 pKa = 5.23VLALAMSLGTPQTEE111 pKa = 4.22SVLASMDD118 pKa = 3.34SSNLVAVPGSWWSGLHH134 pKa = 6.91FDD136 pKa = 5.03DD137 pKa = 5.65ADD139 pKa = 3.91RR140 pKa = 11.84GLILEE145 pKa = 4.71SGYY148 pKa = 10.71SYY150 pKa = 10.08CTEE153 pKa = 4.13SVIGLDD159 pKa = 3.4WFSEE163 pKa = 4.8EE164 pKa = 4.28YY165 pKa = 8.94GTPEE169 pKa = 3.6TLLTVGYY176 pKa = 8.97PGDD179 pKa = 3.97YY180 pKa = 10.9GSDD183 pKa = 3.41SAVGATRR190 pKa = 11.84WAEE193 pKa = 3.98ANDD196 pKa = 3.67VEE198 pKa = 5.09VLAQIDD204 pKa = 3.89TAPNAMVQNQDD215 pKa = 3.09AAVEE219 pKa = 4.41QILSQQPDD227 pKa = 3.53VVLIATAPGEE237 pKa = 3.95AAEE240 pKa = 4.4IVAKK244 pKa = 10.37AVSRR248 pKa = 11.84GYY250 pKa = 10.61DD251 pKa = 3.33GRR253 pKa = 11.84FLGSVPTWNPRR264 pKa = 11.84LLEE267 pKa = 4.01TAAIGALTARR277 pKa = 11.84YY278 pKa = 8.84NHH280 pKa = 6.11VGPWQGWDD288 pKa = 3.31GDD290 pKa = 4.33SAALSAIRR298 pKa = 11.84EE299 pKa = 4.36SLDD302 pKa = 3.13GRR304 pKa = 11.84LPDD307 pKa = 3.38NQGYY311 pKa = 9.45IFGWVWSYY319 pKa = 10.79PLKK322 pKa = 10.92SLLEE326 pKa = 4.03AAADD330 pKa = 3.94DD331 pKa = 4.91GEE333 pKa = 4.44LSRR336 pKa = 11.84SALRR340 pKa = 11.84AAIDD344 pKa = 3.75GLEE347 pKa = 3.82VDD349 pKa = 4.83YY350 pKa = 11.52EE351 pKa = 4.34GALPNRR357 pKa = 11.84TFGEE361 pKa = 4.33PPAFDD366 pKa = 3.95EE367 pKa = 4.4MSATISVPDD376 pKa = 3.84AEE378 pKa = 4.73VEE380 pKa = 4.06LGTRR384 pKa = 11.84AIADD388 pKa = 3.59GVTATTEE395 pKa = 3.95VEE397 pKa = 3.85YY398 pKa = 10.51DD399 pKa = 3.81APCVQPP405 pKa = 4.64
MM1 pKa = 8.14DD2 pKa = 5.57DD3 pKa = 4.44EE4 pKa = 5.0EE5 pKa = 5.96DD6 pKa = 3.46VTAGVGVTEE15 pKa = 4.92EE16 pKa = 4.3PCPDD20 pKa = 4.05AVNADD25 pKa = 3.62NGCIYY30 pKa = 10.74LGVLSDD36 pKa = 4.45LEE38 pKa = 4.39GGPFAEE44 pKa = 5.77LGAQIQQGQLAFWRR58 pKa = 11.84SVNEE62 pKa = 4.11DD63 pKa = 3.29GGIAGYY69 pKa = 10.49DD70 pKa = 3.46VDD72 pKa = 3.15ISRR75 pKa = 11.84YY76 pKa = 6.69TRR78 pKa = 11.84NTAYY82 pKa = 10.63SVDD85 pKa = 3.55QHH87 pKa = 5.26ATEE90 pKa = 4.15YY91 pKa = 10.96QDD93 pKa = 4.28IEE95 pKa = 4.31PHH97 pKa = 5.23VLALAMSLGTPQTEE111 pKa = 4.22SVLASMDD118 pKa = 3.34SSNLVAVPGSWWSGLHH134 pKa = 6.91FDD136 pKa = 5.03DD137 pKa = 5.65ADD139 pKa = 3.91RR140 pKa = 11.84GLILEE145 pKa = 4.71SGYY148 pKa = 10.71SYY150 pKa = 10.08CTEE153 pKa = 4.13SVIGLDD159 pKa = 3.4WFSEE163 pKa = 4.8EE164 pKa = 4.28YY165 pKa = 8.94GTPEE169 pKa = 3.6TLLTVGYY176 pKa = 8.97PGDD179 pKa = 3.97YY180 pKa = 10.9GSDD183 pKa = 3.41SAVGATRR190 pKa = 11.84WAEE193 pKa = 3.98ANDD196 pKa = 3.67VEE198 pKa = 5.09VLAQIDD204 pKa = 3.89TAPNAMVQNQDD215 pKa = 3.09AAVEE219 pKa = 4.41QILSQQPDD227 pKa = 3.53VVLIATAPGEE237 pKa = 3.95AAEE240 pKa = 4.4IVAKK244 pKa = 10.37AVSRR248 pKa = 11.84GYY250 pKa = 10.61DD251 pKa = 3.33GRR253 pKa = 11.84FLGSVPTWNPRR264 pKa = 11.84LLEE267 pKa = 4.01TAAIGALTARR277 pKa = 11.84YY278 pKa = 8.84NHH280 pKa = 6.11VGPWQGWDD288 pKa = 3.31GDD290 pKa = 4.33SAALSAIRR298 pKa = 11.84EE299 pKa = 4.36SLDD302 pKa = 3.13GRR304 pKa = 11.84LPDD307 pKa = 3.38NQGYY311 pKa = 9.45IFGWVWSYY319 pKa = 10.79PLKK322 pKa = 10.92SLLEE326 pKa = 4.03AAADD330 pKa = 3.94DD331 pKa = 4.91GEE333 pKa = 4.44LSRR336 pKa = 11.84SALRR340 pKa = 11.84AAIDD344 pKa = 3.75GLEE347 pKa = 3.82VDD349 pKa = 4.83YY350 pKa = 11.52EE351 pKa = 4.34GALPNRR357 pKa = 11.84TFGEE361 pKa = 4.33PPAFDD366 pKa = 3.95EE367 pKa = 4.4MSATISVPDD376 pKa = 3.84AEE378 pKa = 4.73VEE380 pKa = 4.06LGTRR384 pKa = 11.84AIADD388 pKa = 3.59GVTATTEE395 pKa = 3.95VEE397 pKa = 3.85YY398 pKa = 10.51DD399 pKa = 3.81APCVQPP405 pKa = 4.64
Molecular weight: 43.17 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6EPG9|F6EPG9_HOYSD Binding-protein-dependent transport systems inner membrane component OS=Hoyosella subflava (strain DSM 45089 / JCM 17490 / NBRC 109087 / DQS3-9A1) OX=443218 GN=AS9A_0950 PE=3 SV=1
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84SIISARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.56GRR42 pKa = 11.84AEE44 pKa = 3.81LAVV47 pKa = 3.46
MM1 pKa = 7.69AKK3 pKa = 10.06GKK5 pKa = 8.69RR6 pKa = 11.84TFQPNNRR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84ARR17 pKa = 11.84VHH19 pKa = 5.99GFRR22 pKa = 11.84LRR24 pKa = 11.84MRR26 pKa = 11.84TRR28 pKa = 11.84AGRR31 pKa = 11.84SIISARR37 pKa = 11.84RR38 pKa = 11.84RR39 pKa = 11.84KK40 pKa = 9.56GRR42 pKa = 11.84AEE44 pKa = 3.81LAVV47 pKa = 3.46
Molecular weight: 5.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1471282 |
37 |
7481 |
312.8 |
33.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.481 ± 0.048 | 0.78 ± 0.012 |
5.933 ± 0.03 | 5.903 ± 0.034 |
3.188 ± 0.022 | 8.693 ± 0.046 |
2.265 ± 0.019 | 4.671 ± 0.027 |
2.283 ± 0.029 | 9.97 ± 0.033 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.055 ± 0.016 | 2.23 ± 0.018 |
5.461 ± 0.029 | 3.002 ± 0.02 |
7.275 ± 0.043 | 5.877 ± 0.023 |
5.938 ± 0.022 | 8.479 ± 0.033 |
1.435 ± 0.016 | 2.08 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
