
African horse sickness virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Orbivirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
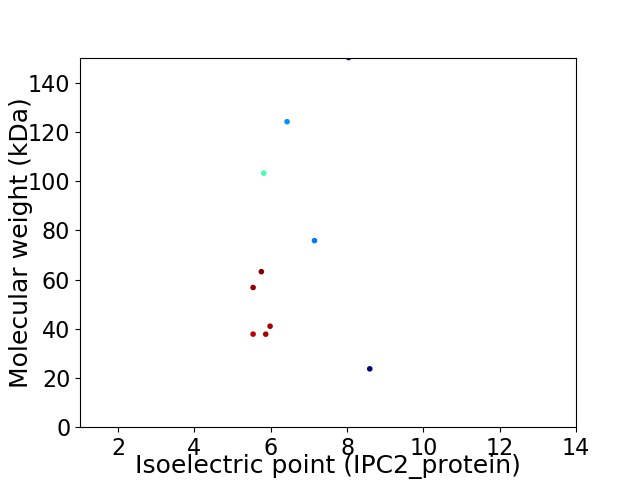
Virtual 2D-PAGE plot for 10 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
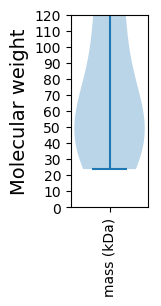
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|D6R1D9|D6R1D9_9REOV Non-structural protein NS1 OS=African horse sickness virus OX=40050 PE=4 SV=1
MM1 pKa = 7.73DD2 pKa = 6.41AIAARR7 pKa = 11.84ALSVVRR13 pKa = 11.84ACVTVTDD20 pKa = 4.54ARR22 pKa = 11.84VSLDD26 pKa = 3.45PGVMEE31 pKa = 4.58TLGIAINRR39 pKa = 11.84YY40 pKa = 9.5NGLTNHH46 pKa = 5.97SVSMRR51 pKa = 11.84PQTQAEE57 pKa = 4.13RR58 pKa = 11.84NEE60 pKa = 4.13MFFMCTDD67 pKa = 3.34MVLAALNVQIGNISPDD83 pKa = 3.27YY84 pKa = 10.33DD85 pKa = 3.29QALATVGALATTEE98 pKa = 3.61IPYY101 pKa = 10.23NVQAMNDD108 pKa = 3.14IVRR111 pKa = 11.84ITGQMQTFGPSKK123 pKa = 10.5VQTGPYY129 pKa = 9.02AGAVEE134 pKa = 4.57VQQSGRR140 pKa = 11.84YY141 pKa = 7.76YY142 pKa = 10.0VPQGRR147 pKa = 11.84TRR149 pKa = 11.84GGYY152 pKa = 9.08INSNIAEE159 pKa = 4.2VCMDD163 pKa = 4.42AGAAGQVNALLAPRR177 pKa = 11.84RR178 pKa = 11.84GDD180 pKa = 2.91AVMIYY185 pKa = 8.43FVWRR189 pKa = 11.84PLRR192 pKa = 11.84IFCDD196 pKa = 3.62PQGASLEE203 pKa = 4.41SAPGTFVTVDD213 pKa = 3.69GVNVAAGDD221 pKa = 3.79VVAWNTIAPVNVGNPGARR239 pKa = 11.84RR240 pKa = 11.84SILQFEE246 pKa = 4.69VLWYY250 pKa = 9.47TSLDD254 pKa = 3.49RR255 pKa = 11.84SLDD258 pKa = 3.83TVPEE262 pKa = 4.24LAPTLTRR269 pKa = 11.84CYY271 pKa = 10.57AYY273 pKa = 10.75VSPTWHH279 pKa = 7.46ALRR282 pKa = 11.84AVIFQQMNMQPINPPIFPPTEE303 pKa = 3.78RR304 pKa = 11.84NEE306 pKa = 3.78IVAYY310 pKa = 10.61LLVASLADD318 pKa = 3.29VYY320 pKa = 11.39AALRR324 pKa = 11.84PDD326 pKa = 3.02FRR328 pKa = 11.84MNGVVAPVGQINRR341 pKa = 11.84ALVLAAYY348 pKa = 8.87HH349 pKa = 6.25
MM1 pKa = 7.73DD2 pKa = 6.41AIAARR7 pKa = 11.84ALSVVRR13 pKa = 11.84ACVTVTDD20 pKa = 4.54ARR22 pKa = 11.84VSLDD26 pKa = 3.45PGVMEE31 pKa = 4.58TLGIAINRR39 pKa = 11.84YY40 pKa = 9.5NGLTNHH46 pKa = 5.97SVSMRR51 pKa = 11.84PQTQAEE57 pKa = 4.13RR58 pKa = 11.84NEE60 pKa = 4.13MFFMCTDD67 pKa = 3.34MVLAALNVQIGNISPDD83 pKa = 3.27YY84 pKa = 10.33DD85 pKa = 3.29QALATVGALATTEE98 pKa = 3.61IPYY101 pKa = 10.23NVQAMNDD108 pKa = 3.14IVRR111 pKa = 11.84ITGQMQTFGPSKK123 pKa = 10.5VQTGPYY129 pKa = 9.02AGAVEE134 pKa = 4.57VQQSGRR140 pKa = 11.84YY141 pKa = 7.76YY142 pKa = 10.0VPQGRR147 pKa = 11.84TRR149 pKa = 11.84GGYY152 pKa = 9.08INSNIAEE159 pKa = 4.2VCMDD163 pKa = 4.42AGAAGQVNALLAPRR177 pKa = 11.84RR178 pKa = 11.84GDD180 pKa = 2.91AVMIYY185 pKa = 8.43FVWRR189 pKa = 11.84PLRR192 pKa = 11.84IFCDD196 pKa = 3.62PQGASLEE203 pKa = 4.41SAPGTFVTVDD213 pKa = 3.69GVNVAAGDD221 pKa = 3.79VVAWNTIAPVNVGNPGARR239 pKa = 11.84RR240 pKa = 11.84SILQFEE246 pKa = 4.69VLWYY250 pKa = 9.47TSLDD254 pKa = 3.49RR255 pKa = 11.84SLDD258 pKa = 3.83TVPEE262 pKa = 4.24LAPTLTRR269 pKa = 11.84CYY271 pKa = 10.57AYY273 pKa = 10.75VSPTWHH279 pKa = 7.46ALRR282 pKa = 11.84AVIFQQMNMQPINPPIFPPTEE303 pKa = 3.78RR304 pKa = 11.84NEE306 pKa = 3.78IVAYY310 pKa = 10.61LLVASLADD318 pKa = 3.29VYY320 pKa = 11.39AALRR324 pKa = 11.84PDD326 pKa = 3.02FRR328 pKa = 11.84MNGVVAPVGQINRR341 pKa = 11.84ALVLAAYY348 pKa = 8.87HH349 pKa = 6.25
Molecular weight: 37.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B4UUP0|B4UUP0_9REOV Outer capsid protein VP5 OS=African horse sickness virus OX=40050 GN=VP5 PE=3 SV=1
MM1 pKa = 7.32NLATIAKK8 pKa = 8.79NYY10 pKa = 10.04SMHH13 pKa = 6.55NGEE16 pKa = 4.51SGAIVPYY23 pKa = 10.42VPPPYY28 pKa = 10.35NFASAPTFSQRR39 pKa = 11.84TSQMEE44 pKa = 4.39SVSLGILNQAMSSTTGASGALKK66 pKa = 10.4DD67 pKa = 3.76EE68 pKa = 4.49KK69 pKa = 11.14AAFGAMAEE77 pKa = 4.23ALRR80 pKa = 11.84DD81 pKa = 3.88PEE83 pKa = 5.29PIRR86 pKa = 11.84QIKK89 pKa = 8.76KK90 pKa = 8.66QVGIRR95 pKa = 11.84TLKK98 pKa = 8.46NLKK101 pKa = 9.25MEE103 pKa = 5.0LATMRR108 pKa = 11.84RR109 pKa = 11.84KK110 pKa = 10.13KK111 pKa = 10.43SALKK115 pKa = 10.48IMIFISGCVTLATSMVGGLSIVDD138 pKa = 3.81DD139 pKa = 5.11EE140 pKa = 5.11ILRR143 pKa = 11.84DD144 pKa = 3.75YY145 pKa = 11.3KK146 pKa = 11.53NNDD149 pKa = 2.78WLMKK153 pKa = 9.64TIHH156 pKa = 6.52GLNLLCTTVLLAAGKK171 pKa = 10.19ISDD174 pKa = 4.01KK175 pKa = 9.65MQEE178 pKa = 4.12EE179 pKa = 4.24ISRR182 pKa = 11.84TKK184 pKa = 10.41RR185 pKa = 11.84DD186 pKa = 3.02IAKK189 pKa = 9.99RR190 pKa = 11.84EE191 pKa = 4.34SYY193 pKa = 11.01VSAASMSWSGDD204 pKa = 3.09TEE206 pKa = 4.02MLLQGIKK213 pKa = 10.53YY214 pKa = 10.44GEE216 pKa = 4.17SS217 pKa = 3.11
MM1 pKa = 7.32NLATIAKK8 pKa = 8.79NYY10 pKa = 10.04SMHH13 pKa = 6.55NGEE16 pKa = 4.51SGAIVPYY23 pKa = 10.42VPPPYY28 pKa = 10.35NFASAPTFSQRR39 pKa = 11.84TSQMEE44 pKa = 4.39SVSLGILNQAMSSTTGASGALKK66 pKa = 10.4DD67 pKa = 3.76EE68 pKa = 4.49KK69 pKa = 11.14AAFGAMAEE77 pKa = 4.23ALRR80 pKa = 11.84DD81 pKa = 3.88PEE83 pKa = 5.29PIRR86 pKa = 11.84QIKK89 pKa = 8.76KK90 pKa = 8.66QVGIRR95 pKa = 11.84TLKK98 pKa = 8.46NLKK101 pKa = 9.25MEE103 pKa = 5.0LATMRR108 pKa = 11.84RR109 pKa = 11.84KK110 pKa = 10.13KK111 pKa = 10.43SALKK115 pKa = 10.48IMIFISGCVTLATSMVGGLSIVDD138 pKa = 3.81DD139 pKa = 5.11EE140 pKa = 5.11ILRR143 pKa = 11.84DD144 pKa = 3.75YY145 pKa = 11.3KK146 pKa = 11.53NNDD149 pKa = 2.78WLMKK153 pKa = 9.64TIHH156 pKa = 6.52GLNLLCTTVLLAAGKK171 pKa = 10.19ISDD174 pKa = 4.01KK175 pKa = 9.65MQEE178 pKa = 4.12EE179 pKa = 4.24ISRR182 pKa = 11.84TKK184 pKa = 10.41RR185 pKa = 11.84DD186 pKa = 3.02IAKK189 pKa = 9.99RR190 pKa = 11.84EE191 pKa = 4.34SYY193 pKa = 11.01VSAASMSWSGDD204 pKa = 3.09TEE206 pKa = 4.02MLLQGIKK213 pKa = 10.53YY214 pKa = 10.44GEE216 pKa = 4.17SS217 pKa = 3.11
Molecular weight: 23.72 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
6262 |
217 |
1305 |
626.2 |
71.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.547 ± 0.878 | 1.214 ± 0.183 |
6.196 ± 0.282 | 7.106 ± 0.676 |
3.881 ± 0.476 | 6.404 ± 0.812 |
1.996 ± 0.24 | 6.771 ± 0.414 |
5.957 ± 0.831 | 8.464 ± 0.322 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.689 ± 0.206 | 4.104 ± 0.356 |
3.801 ± 0.458 | 3.338 ± 0.423 |
7.058 ± 0.291 | 6.052 ± 0.59 |
5.525 ± 0.313 | 6.707 ± 0.356 |
1.262 ± 0.192 | 3.928 ± 0.292 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
