
Perinet vesiculovirus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Rhabdoviridae; Alpharhabdovirinae; Vesiculovirus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
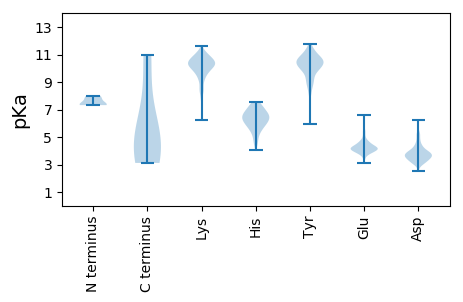
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>tr|I1SV89|I1SV89_9RHAB Matrix protein OS=Perinet vesiculovirus OX=1972569 PE=3 SV=1
MM1 pKa = 7.38SRR3 pKa = 11.84LLEE6 pKa = 4.18TFKK9 pKa = 11.09NYY11 pKa = 9.87PNLEE15 pKa = 3.99KK16 pKa = 10.38TMEE19 pKa = 4.72EE20 pKa = 3.92INDD23 pKa = 3.84IEE25 pKa = 5.14DD26 pKa = 3.48STSQEE31 pKa = 4.09EE32 pKa = 4.48KK33 pKa = 10.7KK34 pKa = 9.33VTLVLSEE41 pKa = 4.1EE42 pKa = 3.97PSYY45 pKa = 11.1FLAHH49 pKa = 6.32MLPSEE54 pKa = 4.04EE55 pKa = 5.41DD56 pKa = 3.33EE57 pKa = 4.21TDD59 pKa = 3.47EE60 pKa = 4.75EE61 pKa = 5.14GGGQVDD67 pKa = 3.64VNPYY71 pKa = 10.11EE72 pKa = 4.43DD73 pKa = 5.23AEE75 pKa = 4.36SSDD78 pKa = 3.67LNEE81 pKa = 5.58DD82 pKa = 3.49EE83 pKa = 5.95FEE85 pKa = 4.7VNFDD89 pKa = 3.78KK90 pKa = 11.28KK91 pKa = 10.32PWTALTEE98 pKa = 4.18KK99 pKa = 10.22VVKK102 pKa = 10.53GNIRR106 pKa = 11.84IDD108 pKa = 3.41MMHH111 pKa = 7.33PEE113 pKa = 4.69GLTDD117 pKa = 3.67AQFTQWVQGIQALLDD132 pKa = 3.6ISHH135 pKa = 6.81HH136 pKa = 6.19VDD138 pKa = 3.74LNRR141 pKa = 11.84LKK143 pKa = 10.83LGQTSDD149 pKa = 4.03GIVLIEE155 pKa = 4.5EE156 pKa = 4.28KK157 pKa = 10.11PKK159 pKa = 10.76RR160 pKa = 11.84PSSPSLKK167 pKa = 10.63LEE169 pKa = 4.1LKK171 pKa = 10.4EE172 pKa = 4.24FVSPDD177 pKa = 3.14SRR179 pKa = 11.84FGTPTDD185 pKa = 3.52NSSEE189 pKa = 4.25SSLGLPPTPEE199 pKa = 3.77KK200 pKa = 11.27VNWKK204 pKa = 7.61QTVEE208 pKa = 4.14IILEE212 pKa = 4.34VKK214 pKa = 10.53ALDD217 pKa = 4.36PDD219 pKa = 3.83LPSYY223 pKa = 9.29KK224 pKa = 8.35TTLSKK229 pKa = 11.12LFGSQEE235 pKa = 3.86AALQYY240 pKa = 10.49CNSGSPMLKK249 pKa = 10.14DD250 pKa = 3.67AFIAGLKK257 pKa = 10.11RR258 pKa = 11.84KK259 pKa = 10.18GIFNRR264 pKa = 11.84MRR266 pKa = 11.84TKK268 pKa = 10.89YY269 pKa = 10.28ILDD272 pKa = 3.59PTLPMM277 pKa = 5.19
MM1 pKa = 7.38SRR3 pKa = 11.84LLEE6 pKa = 4.18TFKK9 pKa = 11.09NYY11 pKa = 9.87PNLEE15 pKa = 3.99KK16 pKa = 10.38TMEE19 pKa = 4.72EE20 pKa = 3.92INDD23 pKa = 3.84IEE25 pKa = 5.14DD26 pKa = 3.48STSQEE31 pKa = 4.09EE32 pKa = 4.48KK33 pKa = 10.7KK34 pKa = 9.33VTLVLSEE41 pKa = 4.1EE42 pKa = 3.97PSYY45 pKa = 11.1FLAHH49 pKa = 6.32MLPSEE54 pKa = 4.04EE55 pKa = 5.41DD56 pKa = 3.33EE57 pKa = 4.21TDD59 pKa = 3.47EE60 pKa = 4.75EE61 pKa = 5.14GGGQVDD67 pKa = 3.64VNPYY71 pKa = 10.11EE72 pKa = 4.43DD73 pKa = 5.23AEE75 pKa = 4.36SSDD78 pKa = 3.67LNEE81 pKa = 5.58DD82 pKa = 3.49EE83 pKa = 5.95FEE85 pKa = 4.7VNFDD89 pKa = 3.78KK90 pKa = 11.28KK91 pKa = 10.32PWTALTEE98 pKa = 4.18KK99 pKa = 10.22VVKK102 pKa = 10.53GNIRR106 pKa = 11.84IDD108 pKa = 3.41MMHH111 pKa = 7.33PEE113 pKa = 4.69GLTDD117 pKa = 3.67AQFTQWVQGIQALLDD132 pKa = 3.6ISHH135 pKa = 6.81HH136 pKa = 6.19VDD138 pKa = 3.74LNRR141 pKa = 11.84LKK143 pKa = 10.83LGQTSDD149 pKa = 4.03GIVLIEE155 pKa = 4.5EE156 pKa = 4.28KK157 pKa = 10.11PKK159 pKa = 10.76RR160 pKa = 11.84PSSPSLKK167 pKa = 10.63LEE169 pKa = 4.1LKK171 pKa = 10.4EE172 pKa = 4.24FVSPDD177 pKa = 3.14SRR179 pKa = 11.84FGTPTDD185 pKa = 3.52NSSEE189 pKa = 4.25SSLGLPPTPEE199 pKa = 3.77KK200 pKa = 11.27VNWKK204 pKa = 7.61QTVEE208 pKa = 4.14IILEE212 pKa = 4.34VKK214 pKa = 10.53ALDD217 pKa = 4.36PDD219 pKa = 3.83LPSYY223 pKa = 9.29KK224 pKa = 8.35TTLSKK229 pKa = 11.12LFGSQEE235 pKa = 3.86AALQYY240 pKa = 10.49CNSGSPMLKK249 pKa = 10.14DD250 pKa = 3.67AFIAGLKK257 pKa = 10.11RR258 pKa = 11.84KK259 pKa = 10.18GIFNRR264 pKa = 11.84MRR266 pKa = 11.84TKK268 pKa = 10.89YY269 pKa = 10.28ILDD272 pKa = 3.59PTLPMM277 pKa = 5.19
Molecular weight: 31.26 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|I1SV90|I1SV90_9RHAB Glycoprotein OS=Perinet vesiculovirus OX=1972569 PE=3 SV=1
MM1 pKa = 7.32NRR3 pKa = 11.84IRR5 pKa = 11.84KK6 pKa = 8.49FWGSNKK12 pKa = 9.65NSQSPEE18 pKa = 3.86KK19 pKa = 10.28KK20 pKa = 9.91SKK22 pKa = 10.37KK23 pKa = 9.51KK24 pKa = 10.41KK25 pKa = 9.93KK26 pKa = 9.91NNNNHH31 pKa = 5.6GSQMEE36 pKa = 4.31WDD38 pKa = 3.88NPPTYY43 pKa = 10.73SEE45 pKa = 4.13AAKK48 pKa = 9.93FDD50 pKa = 3.71PSAPFFGLEE59 pKa = 4.03PEE61 pKa = 4.44DD62 pKa = 3.84MEE64 pKa = 4.52FNYY67 pKa = 10.64YY68 pKa = 10.43SGLASVKK75 pKa = 10.12LQYY78 pKa = 10.89KK79 pKa = 8.38CTIQVRR85 pKa = 11.84AKK87 pKa = 10.08KK88 pKa = 10.03PFVSFLDD95 pKa = 3.56AAEE98 pKa = 4.67NLSAWEE104 pKa = 3.92NNYY107 pKa = 10.78KK108 pKa = 10.7GFLGKK113 pKa = 10.36KK114 pKa = 7.74PFYY117 pKa = 10.13RR118 pKa = 11.84AIILTTLKK126 pKa = 10.27KK127 pKa = 10.73LKK129 pKa = 10.28VSPHH133 pKa = 6.3SLMDD137 pKa = 4.07GNAPEE142 pKa = 4.28YY143 pKa = 10.58NGEE146 pKa = 4.18SEE148 pKa = 4.42GRR150 pKa = 11.84CLILHH155 pKa = 6.23SLGIIPPMMYY165 pKa = 10.06VPEE168 pKa = 4.07QFTRR172 pKa = 11.84EE173 pKa = 3.47WSTRR177 pKa = 11.84DD178 pKa = 3.21NQGIISVRR186 pKa = 11.84ISVGITDD193 pKa = 3.7TVDD196 pKa = 3.1NLEE199 pKa = 4.6GLFSSPVFLDD209 pKa = 3.13HH210 pKa = 7.58KK211 pKa = 10.45EE212 pKa = 4.2LITAGRR218 pKa = 11.84MFNLDD223 pKa = 3.58LKK225 pKa = 11.0VSPEE229 pKa = 4.0NGWFISKK236 pKa = 10.23SYY238 pKa = 10.99
MM1 pKa = 7.32NRR3 pKa = 11.84IRR5 pKa = 11.84KK6 pKa = 8.49FWGSNKK12 pKa = 9.65NSQSPEE18 pKa = 3.86KK19 pKa = 10.28KK20 pKa = 9.91SKK22 pKa = 10.37KK23 pKa = 9.51KK24 pKa = 10.41KK25 pKa = 9.93KK26 pKa = 9.91NNNNHH31 pKa = 5.6GSQMEE36 pKa = 4.31WDD38 pKa = 3.88NPPTYY43 pKa = 10.73SEE45 pKa = 4.13AAKK48 pKa = 9.93FDD50 pKa = 3.71PSAPFFGLEE59 pKa = 4.03PEE61 pKa = 4.44DD62 pKa = 3.84MEE64 pKa = 4.52FNYY67 pKa = 10.64YY68 pKa = 10.43SGLASVKK75 pKa = 10.12LQYY78 pKa = 10.89KK79 pKa = 8.38CTIQVRR85 pKa = 11.84AKK87 pKa = 10.08KK88 pKa = 10.03PFVSFLDD95 pKa = 3.56AAEE98 pKa = 4.67NLSAWEE104 pKa = 3.92NNYY107 pKa = 10.78KK108 pKa = 10.7GFLGKK113 pKa = 10.36KK114 pKa = 7.74PFYY117 pKa = 10.13RR118 pKa = 11.84AIILTTLKK126 pKa = 10.27KK127 pKa = 10.73LKK129 pKa = 10.28VSPHH133 pKa = 6.3SLMDD137 pKa = 4.07GNAPEE142 pKa = 4.28YY143 pKa = 10.58NGEE146 pKa = 4.18SEE148 pKa = 4.42GRR150 pKa = 11.84CLILHH155 pKa = 6.23SLGIIPPMMYY165 pKa = 10.06VPEE168 pKa = 4.07QFTRR172 pKa = 11.84EE173 pKa = 3.47WSTRR177 pKa = 11.84DD178 pKa = 3.21NQGIISVRR186 pKa = 11.84ISVGITDD193 pKa = 3.7TVDD196 pKa = 3.1NLEE199 pKa = 4.6GLFSSPVFLDD209 pKa = 3.13HH210 pKa = 7.58KK211 pKa = 10.45EE212 pKa = 4.2LITAGRR218 pKa = 11.84MFNLDD223 pKa = 3.58LKK225 pKa = 11.0VSPEE229 pKa = 4.0NGWFISKK236 pKa = 10.23SYY238 pKa = 10.99
Molecular weight: 27.16 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3458 |
238 |
2094 |
691.6 |
78.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.714 ± 0.765 | 1.764 ± 0.418 |
6.044 ± 0.525 | 5.726 ± 0.826 |
4.309 ± 0.255 | 5.87 ± 0.293 |
2.4 ± 0.247 | 6.449 ± 0.605 |
6.536 ± 0.565 | 9.919 ± 0.721 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.718 ± 0.407 | 4.685 ± 0.467 |
4.829 ± 0.852 | 3.21 ± 0.128 |
5.35 ± 0.742 | 8.618 ± 0.924 |
5.986 ± 0.588 | 5.552 ± 0.307 |
1.909 ± 0.189 | 3.412 ± 0.355 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
