
Wenzhou bivalvia virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 7.34
Get precalculated fractions of proteins
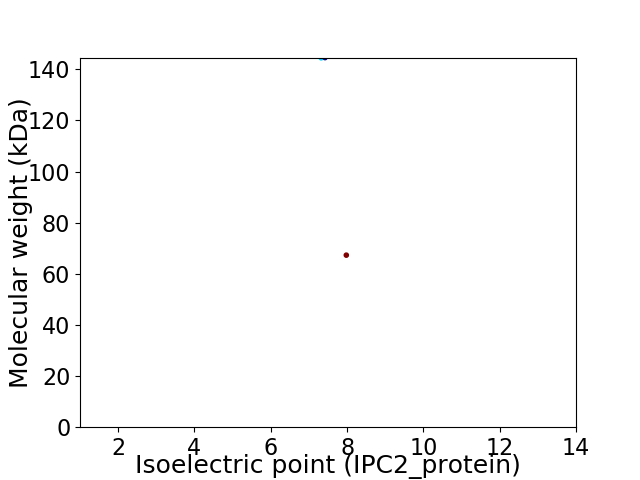
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
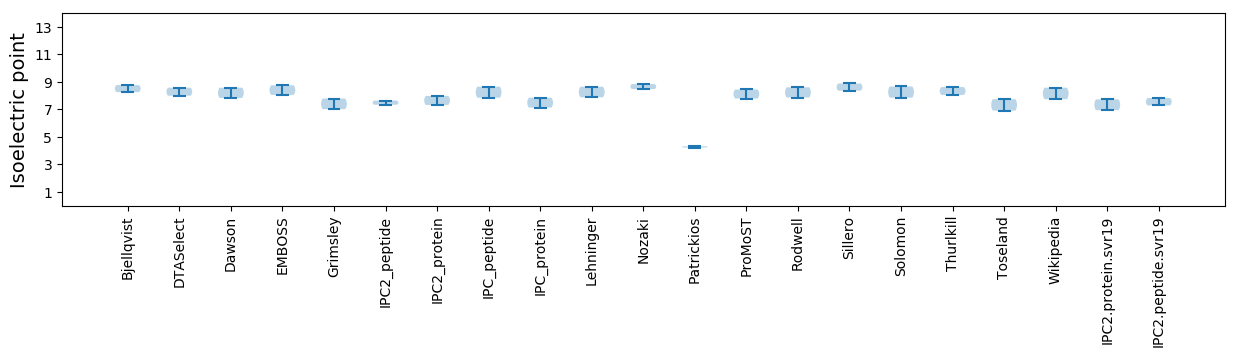
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KL44|A0A1L3KL44_9VIRU Uncharacterized protein OS=Wenzhou bivalvia virus 2 OX=1923554 PE=4 SV=1
MM1 pKa = 7.63TDD3 pKa = 3.17AKK5 pKa = 10.5VKK7 pKa = 10.51RR8 pKa = 11.84PAGRR12 pKa = 11.84QCNNWQPVSHH22 pKa = 6.55PRR24 pKa = 11.84KK25 pKa = 10.01GKK27 pKa = 8.94GHH29 pKa = 5.27VANRR33 pKa = 11.84SNSGLSRR40 pKa = 11.84FYY42 pKa = 11.09CGIKK46 pKa = 10.48GEE48 pKa = 4.24NEE50 pKa = 3.39ANIAVEE56 pKa = 4.64TISLLDD62 pKa = 4.52QINLDD67 pKa = 3.22WKK69 pKa = 10.83SVVKK73 pKa = 10.45RR74 pKa = 11.84HH75 pKa = 6.04ALPDD79 pKa = 4.54GYY81 pKa = 9.98PCALVAFAACLPFEE95 pKa = 3.97EE96 pKa = 5.95HH97 pKa = 6.62IEE99 pKa = 4.02YY100 pKa = 9.91FYY102 pKa = 10.78KK103 pKa = 10.57QLMLNGLLARR113 pKa = 11.84NEE115 pKa = 3.99QGPPGGMSLVGMLKK129 pKa = 10.48ALNTWEE135 pKa = 3.75QAVRR139 pKa = 11.84NKK141 pKa = 10.35RR142 pKa = 11.84FRR144 pKa = 11.84TDD146 pKa = 2.68KK147 pKa = 10.85LRR149 pKa = 11.84GMDD152 pKa = 3.4VHH154 pKa = 7.08HH155 pKa = 6.78ITLKK159 pKa = 10.03GSKK162 pKa = 9.61IVDD165 pKa = 3.89FSIYY169 pKa = 8.62PTGLKK174 pKa = 10.12GSNVEE179 pKa = 4.12TIIIFIEE186 pKa = 4.24GDD188 pKa = 3.09GRR190 pKa = 11.84EE191 pKa = 4.12GHH193 pKa = 6.23FFSANKK199 pKa = 8.2MWPGQHH205 pKa = 6.01YY206 pKa = 10.46RR207 pKa = 11.84MKK209 pKa = 10.62QLAEE213 pKa = 3.8QNRR216 pKa = 11.84LIEE219 pKa = 4.3EE220 pKa = 4.53AAQTTSNVPTPSSEE234 pKa = 3.72PRR236 pKa = 11.84VTVTTPEE243 pKa = 3.84SRR245 pKa = 11.84DD246 pKa = 3.29IEE248 pKa = 4.6TVTWLGLTPDD258 pKa = 3.69EE259 pKa = 4.55LNIVEE264 pKa = 4.59ASVGDD269 pKa = 3.87MDD271 pKa = 5.33PEE273 pKa = 4.46PEE275 pKa = 3.87EE276 pKa = 4.11SRR278 pKa = 11.84EE279 pKa = 4.0EE280 pKa = 3.86IAGQGIMIEE289 pKa = 4.32PYY291 pKa = 10.4EE292 pKa = 4.37PYY294 pKa = 10.18HH295 pKa = 7.32LNGKK299 pKa = 8.92VINTASDD306 pKa = 3.98DD307 pKa = 4.22LPSLIPGQPAMPRR320 pKa = 11.84PRR322 pKa = 11.84VGRR325 pKa = 11.84RR326 pKa = 11.84LSYY329 pKa = 10.92DD330 pKa = 2.56GTMVVWSPDD339 pKa = 3.5PPDD342 pKa = 4.55PPSPPPEE349 pKa = 4.53EE350 pKa = 4.16IRR352 pKa = 11.84PPSPPPSPMLEE363 pKa = 3.98FVPDD367 pKa = 3.53EE368 pKa = 4.44SIEE371 pKa = 4.44FIKK374 pKa = 10.4TNVTVPRR381 pKa = 11.84FTHH384 pKa = 6.74ALRR387 pKa = 11.84GYY389 pKa = 10.74AVGDD393 pKa = 4.09LLPDD397 pKa = 3.9GAVGHH402 pKa = 6.82TDD404 pKa = 3.68ASCEE408 pKa = 4.12NACGATEE415 pKa = 3.88LHH417 pKa = 6.81PAVQEE422 pKa = 4.06KK423 pKa = 10.62FGFTRR428 pKa = 11.84RR429 pKa = 11.84SLYY432 pKa = 10.24YY433 pKa = 9.2VSADD437 pKa = 3.09AGVNPLDD444 pKa = 4.83AKK446 pKa = 11.07YY447 pKa = 8.86LTNGQYY453 pKa = 10.53NPAWLTSMVFGNTSYY468 pKa = 10.83VLEE471 pKa = 4.19YY472 pKa = 11.0VGIFSDD478 pKa = 3.71CHH480 pKa = 5.47FFRR483 pKa = 11.84LAEE486 pKa = 4.01TATIVPKK493 pKa = 10.4SRR495 pKa = 11.84KK496 pKa = 8.69FFSRR500 pKa = 11.84LFSGLGAPAVGTLVCKK516 pKa = 10.37GAALLKK522 pKa = 10.12VLGVAASVGAVGTVSGPIIATAGLAYY548 pKa = 10.15AAFPDD553 pKa = 4.05AVPTLTHH560 pKa = 6.51RR561 pKa = 11.84LVGVLRR567 pKa = 11.84EE568 pKa = 3.82RR569 pKa = 11.84GFISEE574 pKa = 4.89LPAKK578 pKa = 10.08CFHH581 pKa = 6.4AQKK584 pKa = 10.46KK585 pKa = 9.81LPAADD590 pKa = 4.3LSTFDD595 pKa = 3.19GHH597 pKa = 6.9RR598 pKa = 11.84RR599 pKa = 11.84AEE601 pKa = 4.04WTVMRR606 pKa = 11.84QYY608 pKa = 10.91VRR610 pKa = 11.84EE611 pKa = 4.01DD612 pKa = 3.44LVPAFHH618 pKa = 7.13SLQVRR623 pKa = 11.84NGDD626 pKa = 3.44HH627 pKa = 7.65DD628 pKa = 4.56VDD630 pKa = 4.02PEE632 pKa = 3.99MALEE636 pKa = 4.27GLQTLKK642 pKa = 10.75HH643 pKa = 5.91RR644 pKa = 11.84LEE646 pKa = 4.38TKK648 pKa = 10.27RR649 pKa = 11.84GTRR652 pKa = 11.84GYY654 pKa = 10.98KK655 pKa = 9.61VFQTSGPKK663 pKa = 9.32HH664 pKa = 6.52CKK666 pKa = 9.85SCGKK670 pKa = 9.09PPPPAGVKK678 pKa = 9.39YY679 pKa = 9.05KK680 pKa = 9.71WKK682 pKa = 10.4HH683 pKa = 4.98RR684 pKa = 11.84VCTDD688 pKa = 3.17CDD690 pKa = 3.54KK691 pKa = 11.27TLGLCGAITPMGRR704 pKa = 11.84DD705 pKa = 3.05IQANCAVADD714 pKa = 4.35GPPGRR719 pKa = 11.84VHH721 pKa = 6.93MYY723 pKa = 10.78SSTLPPKK730 pKa = 9.69KK731 pKa = 10.3KK732 pKa = 9.53KK733 pKa = 8.87WEE735 pKa = 4.03QVEE738 pKa = 4.47VPPGAITIRR747 pKa = 11.84ASDD750 pKa = 4.48APWMAGVRR758 pKa = 11.84TMAKK762 pKa = 9.2VLEE765 pKa = 4.31VTKK768 pKa = 10.9EE769 pKa = 4.09DD770 pKa = 3.11IFKK773 pKa = 10.26IDD775 pKa = 3.03TSLEE779 pKa = 3.84KK780 pKa = 10.38RR781 pKa = 11.84RR782 pKa = 11.84QEE784 pKa = 3.99CVLAGIAISGCYY796 pKa = 9.85PMVTQKK802 pKa = 11.27GLYY805 pKa = 10.2SRR807 pKa = 11.84MQALLGRR814 pKa = 11.84AFLKK818 pKa = 10.57KK819 pKa = 9.85PQSCPRR825 pKa = 11.84AWKK828 pKa = 10.47KK829 pKa = 10.17MEE831 pKa = 4.67DD832 pKa = 3.95LKK834 pKa = 11.49HH835 pKa = 6.6LILPKK840 pKa = 10.5GALDD844 pKa = 4.26GPQMDD849 pKa = 3.79VEE851 pKa = 4.35EE852 pKa = 5.58WIASMPGRR860 pKa = 11.84RR861 pKa = 11.84KK862 pKa = 9.71RR863 pKa = 11.84ALKK866 pKa = 10.07RR867 pKa = 11.84AYY869 pKa = 9.23KK870 pKa = 10.32QFMEE874 pKa = 5.7DD875 pKa = 3.67GLILEE880 pKa = 4.86KK881 pKa = 10.94DD882 pKa = 3.53LTFSAFVKK890 pKa = 10.66QEE892 pKa = 3.63LLAAFEE898 pKa = 4.59EE899 pKa = 5.03YY900 pKa = 10.19EE901 pKa = 4.47GPVSKK906 pKa = 10.52EE907 pKa = 3.74LEE909 pKa = 3.91EE910 pKa = 4.44TIARR914 pKa = 11.84MIMAPQDD921 pKa = 3.6KK922 pKa = 9.85AHH924 pKa = 6.62VIAGPVIKK932 pKa = 9.99PKK934 pKa = 10.73LMRR937 pKa = 11.84LKK939 pKa = 10.72DD940 pKa = 3.5HH941 pKa = 6.32WHH943 pKa = 6.17HH944 pKa = 7.2DD945 pKa = 2.81NWLFYY950 pKa = 11.1GATTPKK956 pKa = 10.49NLQSWLDD963 pKa = 3.63KK964 pKa = 11.21AVGICADD971 pKa = 3.78GEE973 pKa = 4.8VFTFWCDD980 pKa = 3.48FSMFDD985 pKa = 4.04CTHH988 pKa = 6.46NEE990 pKa = 3.67YY991 pKa = 11.29SMKK994 pKa = 10.53LIEE997 pKa = 4.56SYY999 pKa = 10.27YY1000 pKa = 10.63SEE1002 pKa = 4.82MGTSPLFKK1010 pKa = 10.02MIIDD1014 pKa = 4.1AWRR1017 pKa = 11.84VPAGTMGEE1025 pKa = 4.24LKK1027 pKa = 10.5FRR1029 pKa = 11.84LHH1031 pKa = 7.04QIMLASGRR1039 pKa = 11.84DD1040 pKa = 3.41DD1041 pKa = 3.63TALMNAMYY1049 pKa = 10.02CGFVMGLAVAAAVRR1063 pKa = 11.84NKK1065 pKa = 10.11PLEE1068 pKa = 4.49DD1069 pKa = 4.33LDD1071 pKa = 4.77SGDD1074 pKa = 3.89ILFAAAYY1081 pKa = 9.44VRR1083 pKa = 11.84ISICGDD1089 pKa = 3.13DD1090 pKa = 3.78TLGFLPKK1097 pKa = 10.43NLWFRR1102 pKa = 11.84RR1103 pKa = 11.84AQIMASIEE1111 pKa = 4.31TNLSRR1116 pKa = 11.84FGLVSKK1122 pKa = 10.45LDD1124 pKa = 3.56CSNYY1128 pKa = 10.15LGSAVYY1134 pKa = 10.57LGMRR1138 pKa = 11.84PYY1140 pKa = 10.75NVPTPFGRR1148 pKa = 11.84QWLWGRR1154 pKa = 11.84TIGRR1158 pKa = 11.84AAYY1161 pKa = 9.69KK1162 pKa = 9.2MGWMLDD1168 pKa = 3.64LSKK1171 pKa = 11.4GDD1173 pKa = 3.67AAAWAYY1179 pKa = 10.22GVADD1183 pKa = 5.56AIARR1187 pKa = 11.84TQPYY1191 pKa = 9.79VPLLSDD1197 pKa = 3.97LAKK1200 pKa = 10.53KK1201 pKa = 10.23VVEE1204 pKa = 4.39LSQGYY1209 pKa = 9.45KK1210 pKa = 8.6RR1211 pKa = 11.84TPVLEE1216 pKa = 4.83DD1217 pKa = 3.7PNKK1220 pKa = 10.24PWTHH1224 pKa = 5.29WTPHH1228 pKa = 5.72EE1229 pKa = 4.16NLGQLTYY1236 pKa = 11.16DD1237 pKa = 4.0DD1238 pKa = 4.04QTLEE1242 pKa = 4.34CLVLSYY1248 pKa = 7.98DD1249 pKa = 3.87TPTHH1253 pKa = 6.46HH1254 pKa = 7.08GASQPVSPTIHH1265 pKa = 7.05DD1266 pKa = 3.47LHH1268 pKa = 7.66RR1269 pKa = 11.84SVRR1272 pKa = 11.84NIQKK1276 pKa = 9.6IDD1278 pKa = 3.5RR1279 pKa = 11.84LPYY1282 pKa = 9.98NLEE1285 pKa = 4.45DD1286 pKa = 3.81YY1287 pKa = 11.18ALQCYY1292 pKa = 9.44CNRR1295 pKa = 11.84DD1296 pKa = 3.4DD1297 pKa = 4.91KK1298 pKa = 11.86
MM1 pKa = 7.63TDD3 pKa = 3.17AKK5 pKa = 10.5VKK7 pKa = 10.51RR8 pKa = 11.84PAGRR12 pKa = 11.84QCNNWQPVSHH22 pKa = 6.55PRR24 pKa = 11.84KK25 pKa = 10.01GKK27 pKa = 8.94GHH29 pKa = 5.27VANRR33 pKa = 11.84SNSGLSRR40 pKa = 11.84FYY42 pKa = 11.09CGIKK46 pKa = 10.48GEE48 pKa = 4.24NEE50 pKa = 3.39ANIAVEE56 pKa = 4.64TISLLDD62 pKa = 4.52QINLDD67 pKa = 3.22WKK69 pKa = 10.83SVVKK73 pKa = 10.45RR74 pKa = 11.84HH75 pKa = 6.04ALPDD79 pKa = 4.54GYY81 pKa = 9.98PCALVAFAACLPFEE95 pKa = 3.97EE96 pKa = 5.95HH97 pKa = 6.62IEE99 pKa = 4.02YY100 pKa = 9.91FYY102 pKa = 10.78KK103 pKa = 10.57QLMLNGLLARR113 pKa = 11.84NEE115 pKa = 3.99QGPPGGMSLVGMLKK129 pKa = 10.48ALNTWEE135 pKa = 3.75QAVRR139 pKa = 11.84NKK141 pKa = 10.35RR142 pKa = 11.84FRR144 pKa = 11.84TDD146 pKa = 2.68KK147 pKa = 10.85LRR149 pKa = 11.84GMDD152 pKa = 3.4VHH154 pKa = 7.08HH155 pKa = 6.78ITLKK159 pKa = 10.03GSKK162 pKa = 9.61IVDD165 pKa = 3.89FSIYY169 pKa = 8.62PTGLKK174 pKa = 10.12GSNVEE179 pKa = 4.12TIIIFIEE186 pKa = 4.24GDD188 pKa = 3.09GRR190 pKa = 11.84EE191 pKa = 4.12GHH193 pKa = 6.23FFSANKK199 pKa = 8.2MWPGQHH205 pKa = 6.01YY206 pKa = 10.46RR207 pKa = 11.84MKK209 pKa = 10.62QLAEE213 pKa = 3.8QNRR216 pKa = 11.84LIEE219 pKa = 4.3EE220 pKa = 4.53AAQTTSNVPTPSSEE234 pKa = 3.72PRR236 pKa = 11.84VTVTTPEE243 pKa = 3.84SRR245 pKa = 11.84DD246 pKa = 3.29IEE248 pKa = 4.6TVTWLGLTPDD258 pKa = 3.69EE259 pKa = 4.55LNIVEE264 pKa = 4.59ASVGDD269 pKa = 3.87MDD271 pKa = 5.33PEE273 pKa = 4.46PEE275 pKa = 3.87EE276 pKa = 4.11SRR278 pKa = 11.84EE279 pKa = 4.0EE280 pKa = 3.86IAGQGIMIEE289 pKa = 4.32PYY291 pKa = 10.4EE292 pKa = 4.37PYY294 pKa = 10.18HH295 pKa = 7.32LNGKK299 pKa = 8.92VINTASDD306 pKa = 3.98DD307 pKa = 4.22LPSLIPGQPAMPRR320 pKa = 11.84PRR322 pKa = 11.84VGRR325 pKa = 11.84RR326 pKa = 11.84LSYY329 pKa = 10.92DD330 pKa = 2.56GTMVVWSPDD339 pKa = 3.5PPDD342 pKa = 4.55PPSPPPEE349 pKa = 4.53EE350 pKa = 4.16IRR352 pKa = 11.84PPSPPPSPMLEE363 pKa = 3.98FVPDD367 pKa = 3.53EE368 pKa = 4.44SIEE371 pKa = 4.44FIKK374 pKa = 10.4TNVTVPRR381 pKa = 11.84FTHH384 pKa = 6.74ALRR387 pKa = 11.84GYY389 pKa = 10.74AVGDD393 pKa = 4.09LLPDD397 pKa = 3.9GAVGHH402 pKa = 6.82TDD404 pKa = 3.68ASCEE408 pKa = 4.12NACGATEE415 pKa = 3.88LHH417 pKa = 6.81PAVQEE422 pKa = 4.06KK423 pKa = 10.62FGFTRR428 pKa = 11.84RR429 pKa = 11.84SLYY432 pKa = 10.24YY433 pKa = 9.2VSADD437 pKa = 3.09AGVNPLDD444 pKa = 4.83AKK446 pKa = 11.07YY447 pKa = 8.86LTNGQYY453 pKa = 10.53NPAWLTSMVFGNTSYY468 pKa = 10.83VLEE471 pKa = 4.19YY472 pKa = 11.0VGIFSDD478 pKa = 3.71CHH480 pKa = 5.47FFRR483 pKa = 11.84LAEE486 pKa = 4.01TATIVPKK493 pKa = 10.4SRR495 pKa = 11.84KK496 pKa = 8.69FFSRR500 pKa = 11.84LFSGLGAPAVGTLVCKK516 pKa = 10.37GAALLKK522 pKa = 10.12VLGVAASVGAVGTVSGPIIATAGLAYY548 pKa = 10.15AAFPDD553 pKa = 4.05AVPTLTHH560 pKa = 6.51RR561 pKa = 11.84LVGVLRR567 pKa = 11.84EE568 pKa = 3.82RR569 pKa = 11.84GFISEE574 pKa = 4.89LPAKK578 pKa = 10.08CFHH581 pKa = 6.4AQKK584 pKa = 10.46KK585 pKa = 9.81LPAADD590 pKa = 4.3LSTFDD595 pKa = 3.19GHH597 pKa = 6.9RR598 pKa = 11.84RR599 pKa = 11.84AEE601 pKa = 4.04WTVMRR606 pKa = 11.84QYY608 pKa = 10.91VRR610 pKa = 11.84EE611 pKa = 4.01DD612 pKa = 3.44LVPAFHH618 pKa = 7.13SLQVRR623 pKa = 11.84NGDD626 pKa = 3.44HH627 pKa = 7.65DD628 pKa = 4.56VDD630 pKa = 4.02PEE632 pKa = 3.99MALEE636 pKa = 4.27GLQTLKK642 pKa = 10.75HH643 pKa = 5.91RR644 pKa = 11.84LEE646 pKa = 4.38TKK648 pKa = 10.27RR649 pKa = 11.84GTRR652 pKa = 11.84GYY654 pKa = 10.98KK655 pKa = 9.61VFQTSGPKK663 pKa = 9.32HH664 pKa = 6.52CKK666 pKa = 9.85SCGKK670 pKa = 9.09PPPPAGVKK678 pKa = 9.39YY679 pKa = 9.05KK680 pKa = 9.71WKK682 pKa = 10.4HH683 pKa = 4.98RR684 pKa = 11.84VCTDD688 pKa = 3.17CDD690 pKa = 3.54KK691 pKa = 11.27TLGLCGAITPMGRR704 pKa = 11.84DD705 pKa = 3.05IQANCAVADD714 pKa = 4.35GPPGRR719 pKa = 11.84VHH721 pKa = 6.93MYY723 pKa = 10.78SSTLPPKK730 pKa = 9.69KK731 pKa = 10.3KK732 pKa = 9.53KK733 pKa = 8.87WEE735 pKa = 4.03QVEE738 pKa = 4.47VPPGAITIRR747 pKa = 11.84ASDD750 pKa = 4.48APWMAGVRR758 pKa = 11.84TMAKK762 pKa = 9.2VLEE765 pKa = 4.31VTKK768 pKa = 10.9EE769 pKa = 4.09DD770 pKa = 3.11IFKK773 pKa = 10.26IDD775 pKa = 3.03TSLEE779 pKa = 3.84KK780 pKa = 10.38RR781 pKa = 11.84RR782 pKa = 11.84QEE784 pKa = 3.99CVLAGIAISGCYY796 pKa = 9.85PMVTQKK802 pKa = 11.27GLYY805 pKa = 10.2SRR807 pKa = 11.84MQALLGRR814 pKa = 11.84AFLKK818 pKa = 10.57KK819 pKa = 9.85PQSCPRR825 pKa = 11.84AWKK828 pKa = 10.47KK829 pKa = 10.17MEE831 pKa = 4.67DD832 pKa = 3.95LKK834 pKa = 11.49HH835 pKa = 6.6LILPKK840 pKa = 10.5GALDD844 pKa = 4.26GPQMDD849 pKa = 3.79VEE851 pKa = 4.35EE852 pKa = 5.58WIASMPGRR860 pKa = 11.84RR861 pKa = 11.84KK862 pKa = 9.71RR863 pKa = 11.84ALKK866 pKa = 10.07RR867 pKa = 11.84AYY869 pKa = 9.23KK870 pKa = 10.32QFMEE874 pKa = 5.7DD875 pKa = 3.67GLILEE880 pKa = 4.86KK881 pKa = 10.94DD882 pKa = 3.53LTFSAFVKK890 pKa = 10.66QEE892 pKa = 3.63LLAAFEE898 pKa = 4.59EE899 pKa = 5.03YY900 pKa = 10.19EE901 pKa = 4.47GPVSKK906 pKa = 10.52EE907 pKa = 3.74LEE909 pKa = 3.91EE910 pKa = 4.44TIARR914 pKa = 11.84MIMAPQDD921 pKa = 3.6KK922 pKa = 9.85AHH924 pKa = 6.62VIAGPVIKK932 pKa = 9.99PKK934 pKa = 10.73LMRR937 pKa = 11.84LKK939 pKa = 10.72DD940 pKa = 3.5HH941 pKa = 6.32WHH943 pKa = 6.17HH944 pKa = 7.2DD945 pKa = 2.81NWLFYY950 pKa = 11.1GATTPKK956 pKa = 10.49NLQSWLDD963 pKa = 3.63KK964 pKa = 11.21AVGICADD971 pKa = 3.78GEE973 pKa = 4.8VFTFWCDD980 pKa = 3.48FSMFDD985 pKa = 4.04CTHH988 pKa = 6.46NEE990 pKa = 3.67YY991 pKa = 11.29SMKK994 pKa = 10.53LIEE997 pKa = 4.56SYY999 pKa = 10.27YY1000 pKa = 10.63SEE1002 pKa = 4.82MGTSPLFKK1010 pKa = 10.02MIIDD1014 pKa = 4.1AWRR1017 pKa = 11.84VPAGTMGEE1025 pKa = 4.24LKK1027 pKa = 10.5FRR1029 pKa = 11.84LHH1031 pKa = 7.04QIMLASGRR1039 pKa = 11.84DD1040 pKa = 3.41DD1041 pKa = 3.63TALMNAMYY1049 pKa = 10.02CGFVMGLAVAAAVRR1063 pKa = 11.84NKK1065 pKa = 10.11PLEE1068 pKa = 4.49DD1069 pKa = 4.33LDD1071 pKa = 4.77SGDD1074 pKa = 3.89ILFAAAYY1081 pKa = 9.44VRR1083 pKa = 11.84ISICGDD1089 pKa = 3.13DD1090 pKa = 3.78TLGFLPKK1097 pKa = 10.43NLWFRR1102 pKa = 11.84RR1103 pKa = 11.84AQIMASIEE1111 pKa = 4.31TNLSRR1116 pKa = 11.84FGLVSKK1122 pKa = 10.45LDD1124 pKa = 3.56CSNYY1128 pKa = 10.15LGSAVYY1134 pKa = 10.57LGMRR1138 pKa = 11.84PYY1140 pKa = 10.75NVPTPFGRR1148 pKa = 11.84QWLWGRR1154 pKa = 11.84TIGRR1158 pKa = 11.84AAYY1161 pKa = 9.69KK1162 pKa = 9.2MGWMLDD1168 pKa = 3.64LSKK1171 pKa = 11.4GDD1173 pKa = 3.67AAAWAYY1179 pKa = 10.22GVADD1183 pKa = 5.56AIARR1187 pKa = 11.84TQPYY1191 pKa = 9.79VPLLSDD1197 pKa = 3.97LAKK1200 pKa = 10.53KK1201 pKa = 10.23VVEE1204 pKa = 4.39LSQGYY1209 pKa = 9.45KK1210 pKa = 8.6RR1211 pKa = 11.84TPVLEE1216 pKa = 4.83DD1217 pKa = 3.7PNKK1220 pKa = 10.24PWTHH1224 pKa = 5.29WTPHH1228 pKa = 5.72EE1229 pKa = 4.16NLGQLTYY1236 pKa = 11.16DD1237 pKa = 4.0DD1238 pKa = 4.04QTLEE1242 pKa = 4.34CLVLSYY1248 pKa = 7.98DD1249 pKa = 3.87TPTHH1253 pKa = 6.46HH1254 pKa = 7.08GASQPVSPTIHH1265 pKa = 7.05DD1266 pKa = 3.47LHH1268 pKa = 7.66RR1269 pKa = 11.84SVRR1272 pKa = 11.84NIQKK1276 pKa = 9.6IDD1278 pKa = 3.5RR1279 pKa = 11.84LPYY1282 pKa = 9.98NLEE1285 pKa = 4.45DD1286 pKa = 3.81YY1287 pKa = 11.18ALQCYY1292 pKa = 9.44CNRR1295 pKa = 11.84DD1296 pKa = 3.4DD1297 pKa = 4.91KK1298 pKa = 11.86
Molecular weight: 144.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KL44|A0A1L3KL44_9VIRU Uncharacterized protein OS=Wenzhou bivalvia virus 2 OX=1923554 PE=4 SV=1
MM1 pKa = 7.34TFLSAVLLPVRR12 pKa = 11.84AKK14 pKa = 10.43EE15 pKa = 3.63RR16 pKa = 11.84TARR19 pKa = 11.84VMSHH23 pKa = 6.56TGLKK27 pKa = 10.59SLDD30 pKa = 3.37QVAQTICLPNEE41 pKa = 4.01RR42 pKa = 11.84APVRR46 pKa = 11.84LPTYY50 pKa = 10.14PSIDD54 pKa = 3.03KK55 pKa = 9.25TALFRR60 pKa = 11.84YY61 pKa = 9.49RR62 pKa = 11.84YY63 pKa = 10.09QNTEE67 pKa = 3.71SLKK70 pKa = 10.6DD71 pKa = 3.87DD72 pKa = 3.94KK73 pKa = 11.13LVPPGDD79 pKa = 4.76LDD81 pKa = 3.71TLNIPGRR88 pKa = 11.84KK89 pKa = 8.84RR90 pKa = 11.84FILSRR95 pKa = 11.84DD96 pKa = 3.12PAAPMLLDD104 pKa = 3.87SVHH107 pKa = 6.89LLQHH111 pKa = 5.65TWGLDD116 pKa = 3.02PGAFDD121 pKa = 3.22GTIFIRR127 pKa = 11.84GEE129 pKa = 3.87TVYY132 pKa = 10.98LVDD135 pKa = 3.98SASGPSTVHH144 pKa = 6.52SVDD147 pKa = 3.8FDD149 pKa = 3.18KK150 pKa = 11.1CYY152 pKa = 10.1PIEE155 pKa = 4.1YY156 pKa = 10.07YY157 pKa = 8.98RR158 pKa = 11.84TLPAGRR164 pKa = 11.84LDD166 pKa = 3.5GKK168 pKa = 9.76EE169 pKa = 3.5WFMVPRR175 pKa = 11.84VLDD178 pKa = 3.55SRR180 pKa = 11.84GKK182 pKa = 9.38SQPFLNVLCVGLITVDD198 pKa = 4.11GPWPFAANPLGKK210 pKa = 10.44GLIRR214 pKa = 11.84TYY216 pKa = 10.87HH217 pKa = 6.36SDD219 pKa = 3.64ASVTSLNASNGEE231 pKa = 3.87LCLDD235 pKa = 3.45YY236 pKa = 10.77TITIEE241 pKa = 4.74AMSGDD246 pKa = 3.71GAVQEE251 pKa = 5.26LVMSYY256 pKa = 10.8DD257 pKa = 3.08WSNWDD262 pKa = 3.33YY263 pKa = 11.17TGSFDD268 pKa = 4.91LHH270 pKa = 6.84PDD272 pKa = 3.03VAMVRR277 pKa = 11.84IKK279 pKa = 10.83SLTYY283 pKa = 9.29RR284 pKa = 11.84GLVRR288 pKa = 11.84LVDD291 pKa = 3.92NNLPGADD298 pKa = 3.47VPMKK302 pKa = 10.46GCYY305 pKa = 8.71PVLGLSRR312 pKa = 11.84SGAHH316 pKa = 6.22IPAGSPPTTFRR327 pKa = 11.84CLSLPPTAVNPEE339 pKa = 4.36YY340 pKa = 10.21YY341 pKa = 10.6NSVAPFQSTRR351 pKa = 11.84LNSSAVLLTNVSKK364 pKa = 11.05VLNKK368 pKa = 10.29EE369 pKa = 3.84GTVQSSRR376 pKa = 11.84LLFNPNAGQTVHH388 pKa = 6.62HH389 pKa = 7.25ADD391 pKa = 3.38VVSVSTSNPDD401 pKa = 2.8TRR403 pKa = 11.84YY404 pKa = 10.23FGALEE409 pKa = 3.95KK410 pKa = 10.7GAYY413 pKa = 8.23TFTAPDD419 pKa = 3.42QEE421 pKa = 4.44SLKK424 pKa = 10.79FVTPYY429 pKa = 9.55VTVDD433 pKa = 3.23VNDD436 pKa = 4.04AGTGEE441 pKa = 4.24GNIAILTLVSTKK453 pKa = 10.74NVVRR457 pKa = 11.84PVLDD461 pKa = 4.68LGAKK465 pKa = 9.44YY466 pKa = 9.79YY467 pKa = 11.2NCIICTDD474 pKa = 4.35LDD476 pKa = 4.05STDD479 pKa = 3.75DD480 pKa = 3.65TQLALTLDD488 pKa = 3.78THH490 pKa = 5.87WEE492 pKa = 3.82FRR494 pKa = 11.84TISTLYY500 pKa = 8.6TLDD503 pKa = 3.39YY504 pKa = 11.04SRR506 pKa = 11.84IPMEE510 pKa = 4.32VYY512 pKa = 9.64HH513 pKa = 6.86AAMLAVVKK521 pKa = 10.88AGFFYY526 pKa = 10.94EE527 pKa = 4.38NNNHH531 pKa = 6.14AAILRR536 pKa = 11.84MLSAGVKK543 pKa = 9.79FAAPLIAGYY552 pKa = 9.69AARR555 pKa = 11.84IPAGFLQAQATSALAKK571 pKa = 10.22GAVNLALHH579 pKa = 6.38GAKK582 pKa = 9.69KK583 pKa = 10.08AHH585 pKa = 6.21AAYY588 pKa = 7.75RR589 pKa = 11.84THH591 pKa = 6.69QNNKK595 pKa = 8.54QKK597 pKa = 10.57QKK599 pKa = 10.4PKK601 pKa = 10.16NKK603 pKa = 9.16EE604 pKa = 3.75RR605 pKa = 11.84GNMRR609 pKa = 11.84QKK611 pKa = 11.12GLRR614 pKa = 3.51
MM1 pKa = 7.34TFLSAVLLPVRR12 pKa = 11.84AKK14 pKa = 10.43EE15 pKa = 3.63RR16 pKa = 11.84TARR19 pKa = 11.84VMSHH23 pKa = 6.56TGLKK27 pKa = 10.59SLDD30 pKa = 3.37QVAQTICLPNEE41 pKa = 4.01RR42 pKa = 11.84APVRR46 pKa = 11.84LPTYY50 pKa = 10.14PSIDD54 pKa = 3.03KK55 pKa = 9.25TALFRR60 pKa = 11.84YY61 pKa = 9.49RR62 pKa = 11.84YY63 pKa = 10.09QNTEE67 pKa = 3.71SLKK70 pKa = 10.6DD71 pKa = 3.87DD72 pKa = 3.94KK73 pKa = 11.13LVPPGDD79 pKa = 4.76LDD81 pKa = 3.71TLNIPGRR88 pKa = 11.84KK89 pKa = 8.84RR90 pKa = 11.84FILSRR95 pKa = 11.84DD96 pKa = 3.12PAAPMLLDD104 pKa = 3.87SVHH107 pKa = 6.89LLQHH111 pKa = 5.65TWGLDD116 pKa = 3.02PGAFDD121 pKa = 3.22GTIFIRR127 pKa = 11.84GEE129 pKa = 3.87TVYY132 pKa = 10.98LVDD135 pKa = 3.98SASGPSTVHH144 pKa = 6.52SVDD147 pKa = 3.8FDD149 pKa = 3.18KK150 pKa = 11.1CYY152 pKa = 10.1PIEE155 pKa = 4.1YY156 pKa = 10.07YY157 pKa = 8.98RR158 pKa = 11.84TLPAGRR164 pKa = 11.84LDD166 pKa = 3.5GKK168 pKa = 9.76EE169 pKa = 3.5WFMVPRR175 pKa = 11.84VLDD178 pKa = 3.55SRR180 pKa = 11.84GKK182 pKa = 9.38SQPFLNVLCVGLITVDD198 pKa = 4.11GPWPFAANPLGKK210 pKa = 10.44GLIRR214 pKa = 11.84TYY216 pKa = 10.87HH217 pKa = 6.36SDD219 pKa = 3.64ASVTSLNASNGEE231 pKa = 3.87LCLDD235 pKa = 3.45YY236 pKa = 10.77TITIEE241 pKa = 4.74AMSGDD246 pKa = 3.71GAVQEE251 pKa = 5.26LVMSYY256 pKa = 10.8DD257 pKa = 3.08WSNWDD262 pKa = 3.33YY263 pKa = 11.17TGSFDD268 pKa = 4.91LHH270 pKa = 6.84PDD272 pKa = 3.03VAMVRR277 pKa = 11.84IKK279 pKa = 10.83SLTYY283 pKa = 9.29RR284 pKa = 11.84GLVRR288 pKa = 11.84LVDD291 pKa = 3.92NNLPGADD298 pKa = 3.47VPMKK302 pKa = 10.46GCYY305 pKa = 8.71PVLGLSRR312 pKa = 11.84SGAHH316 pKa = 6.22IPAGSPPTTFRR327 pKa = 11.84CLSLPPTAVNPEE339 pKa = 4.36YY340 pKa = 10.21YY341 pKa = 10.6NSVAPFQSTRR351 pKa = 11.84LNSSAVLLTNVSKK364 pKa = 11.05VLNKK368 pKa = 10.29EE369 pKa = 3.84GTVQSSRR376 pKa = 11.84LLFNPNAGQTVHH388 pKa = 6.62HH389 pKa = 7.25ADD391 pKa = 3.38VVSVSTSNPDD401 pKa = 2.8TRR403 pKa = 11.84YY404 pKa = 10.23FGALEE409 pKa = 3.95KK410 pKa = 10.7GAYY413 pKa = 8.23TFTAPDD419 pKa = 3.42QEE421 pKa = 4.44SLKK424 pKa = 10.79FVTPYY429 pKa = 9.55VTVDD433 pKa = 3.23VNDD436 pKa = 4.04AGTGEE441 pKa = 4.24GNIAILTLVSTKK453 pKa = 10.74NVVRR457 pKa = 11.84PVLDD461 pKa = 4.68LGAKK465 pKa = 9.44YY466 pKa = 9.79YY467 pKa = 11.2NCIICTDD474 pKa = 4.35LDD476 pKa = 4.05STDD479 pKa = 3.75DD480 pKa = 3.65TQLALTLDD488 pKa = 3.78THH490 pKa = 5.87WEE492 pKa = 3.82FRR494 pKa = 11.84TISTLYY500 pKa = 8.6TLDD503 pKa = 3.39YY504 pKa = 11.04SRR506 pKa = 11.84IPMEE510 pKa = 4.32VYY512 pKa = 9.64HH513 pKa = 6.86AAMLAVVKK521 pKa = 10.88AGFFYY526 pKa = 10.94EE527 pKa = 4.38NNNHH531 pKa = 6.14AAILRR536 pKa = 11.84MLSAGVKK543 pKa = 9.79FAAPLIAGYY552 pKa = 9.69AARR555 pKa = 11.84IPAGFLQAQATSALAKK571 pKa = 10.22GAVNLALHH579 pKa = 6.38GAKK582 pKa = 9.69KK583 pKa = 10.08AHH585 pKa = 6.21AAYY588 pKa = 7.75RR589 pKa = 11.84THH591 pKa = 6.69QNNKK595 pKa = 8.54QKK597 pKa = 10.57QKK599 pKa = 10.4PKK601 pKa = 10.16NKK603 pKa = 9.16EE604 pKa = 3.75RR605 pKa = 11.84GNMRR609 pKa = 11.84QKK611 pKa = 11.12GLRR614 pKa = 3.51
Molecular weight: 67.31 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1912 |
614 |
1298 |
956.0 |
105.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.682 ± 0.257 | 1.831 ± 0.309 |
5.701 ± 0.191 | 4.759 ± 1.071 |
3.504 ± 0.049 | 7.27 ± 0.347 |
2.615 ± 0.101 | 4.184 ± 0.352 |
5.596 ± 0.512 | 9.623 ± 0.755 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.772 ± 0.479 | 3.661 ± 0.718 |
6.956 ± 0.354 | 2.981 ± 0.124 |
5.596 ± 0.13 | 6.015 ± 0.579 |
6.119 ± 0.804 | 6.904 ± 0.536 |
1.621 ± 0.377 | 3.609 ± 0.367 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
