
Burkholderia phage BcepMu (isolate -/United States/Summer/2002) (Bacteriophage BcepMu)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Myoviridae; Bcepmuvirus; Burkholderia virus BcepMu
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
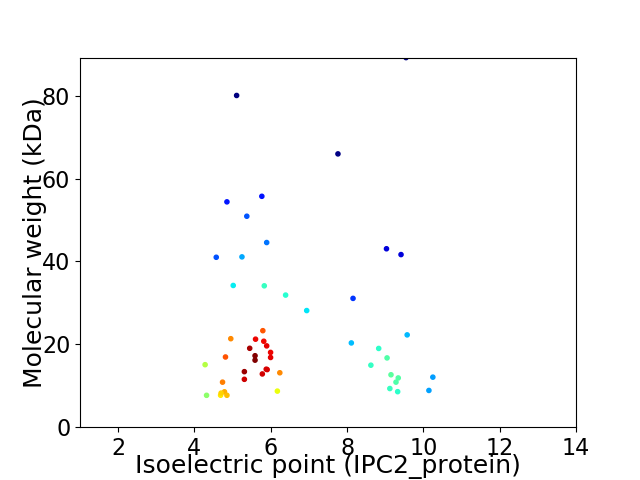
Virtual 2D-PAGE plot for 53 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6QIA4|Q6QIA4_BPBMU Gp45 OS=Burkholderia phage BcepMu (isolate -/United States/Summer/2002) OX=1283335 GN=BcepMu45 PE=4 SV=1
MM1 pKa = 7.51FLTHH5 pKa = 6.41ITTEE9 pKa = 4.19GEE11 pKa = 3.76RR12 pKa = 11.84WDD14 pKa = 3.92QIAYY18 pKa = 9.87RR19 pKa = 11.84YY20 pKa = 10.35YY21 pKa = 10.76GDD23 pKa = 3.47PFAYY27 pKa = 9.81EE28 pKa = 4.49RR29 pKa = 11.84IIAANPSVPITAVLASGIALSIPVVAADD57 pKa = 3.98DD58 pKa = 4.06VADD61 pKa = 4.13EE62 pKa = 4.74EE63 pKa = 4.71LPPWMRR69 pKa = 3.95
MM1 pKa = 7.51FLTHH5 pKa = 6.41ITTEE9 pKa = 4.19GEE11 pKa = 3.76RR12 pKa = 11.84WDD14 pKa = 3.92QIAYY18 pKa = 9.87RR19 pKa = 11.84YY20 pKa = 10.35YY21 pKa = 10.76GDD23 pKa = 3.47PFAYY27 pKa = 9.81EE28 pKa = 4.49RR29 pKa = 11.84IIAANPSVPITAVLASGIALSIPVVAADD57 pKa = 3.98DD58 pKa = 4.06VADD61 pKa = 4.13EE62 pKa = 4.74EE63 pKa = 4.71LPPWMRR69 pKa = 3.95
Molecular weight: 7.7 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6QIA6|Q6QIA6_BPBMU Gp43 (Fragment) OS=Burkholderia phage BcepMu (isolate -/United States/Summer/2002) OX=1283335 GN=BcepMu43 PE=4 SV=1
MM1 pKa = 7.57ASEE4 pKa = 4.93FYY6 pKa = 10.53IGVKK10 pKa = 10.01IGATLLGSFGAALSGTRR27 pKa = 11.84TTLNGLGRR35 pKa = 11.84VADD38 pKa = 4.21EE39 pKa = 3.62LRR41 pKa = 11.84ARR43 pKa = 11.84HH44 pKa = 5.45TRR46 pKa = 11.84LGDD49 pKa = 3.19AMARR53 pKa = 11.84AVAHH57 pKa = 6.07PMRR60 pKa = 11.84NIAEE64 pKa = 4.1LRR66 pKa = 11.84GQYY69 pKa = 10.5DD70 pKa = 3.37RR71 pKa = 11.84LGRR74 pKa = 11.84TIDD77 pKa = 3.44QVQAKK82 pKa = 8.79QAALATRR89 pKa = 11.84LARR92 pKa = 11.84GAALRR97 pKa = 11.84EE98 pKa = 3.92QRR100 pKa = 11.84QGLGADD106 pKa = 3.42MLGTYY111 pKa = 8.0ATAAATAAPVIGAVRR126 pKa = 11.84QAATFEE132 pKa = 4.2AGLRR136 pKa = 11.84DD137 pKa = 3.5IAITGNLTRR146 pKa = 11.84DD147 pKa = 3.09EE148 pKa = 4.22EE149 pKa = 4.22FRR151 pKa = 11.84IGEE154 pKa = 4.27TMRR157 pKa = 11.84RR158 pKa = 11.84AALATSQGHH167 pKa = 5.08NSILEE172 pKa = 4.22GVGTLVAAGMDD183 pKa = 3.48AKK185 pKa = 10.87EE186 pKa = 4.62AGQKK190 pKa = 10.64SNLLGRR196 pKa = 11.84VATATNADD204 pKa = 3.77MKK206 pKa = 10.84DD207 pKa = 3.54LAGMVYY213 pKa = 10.26SFSEE217 pKa = 4.06TLGIKK222 pKa = 10.17GDD224 pKa = 3.84AALKK228 pKa = 9.69EE229 pKa = 4.3AFNRR233 pKa = 11.84AAYY236 pKa = 9.71GGKK239 pKa = 10.07LGRR242 pKa = 11.84FEE244 pKa = 5.05LKK246 pKa = 10.8DD247 pKa = 3.31MAKK250 pKa = 10.47ALPEE254 pKa = 3.77MTAAFAAKK262 pKa = 10.08GIKK265 pKa = 10.08GQDD268 pKa = 3.2ALTQIIASLEE278 pKa = 4.07VGRR281 pKa = 11.84EE282 pKa = 3.94GAGSGDD288 pKa = 3.37EE289 pKa = 4.43AVTNLRR295 pKa = 11.84NWLSHH300 pKa = 5.73MNAKK304 pKa = 8.64ATIDD308 pKa = 3.52AYY310 pKa = 10.11KK311 pKa = 10.38KK312 pKa = 10.77AGVDD316 pKa = 3.73YY317 pKa = 10.29QKK319 pKa = 11.73SMSNLVAGGYY329 pKa = 9.46SSYY332 pKa = 11.01EE333 pKa = 3.64GSLQIAQKK341 pKa = 10.33FIASRR346 pKa = 11.84GDD348 pKa = 3.14AFMKK352 pKa = 9.23QWKK355 pKa = 9.13AAGAKK360 pKa = 9.93GDD362 pKa = 3.88EE363 pKa = 4.14EE364 pKa = 4.25AQRR367 pKa = 11.84KK368 pKa = 8.89LMEE371 pKa = 4.45SFGLNEE377 pKa = 4.17VFQDD381 pKa = 3.12IQTINHH387 pKa = 7.1LLAMRR392 pKa = 11.84QGWDD396 pKa = 3.12KK397 pKa = 11.1YY398 pKa = 7.82QQNKK402 pKa = 9.53KK403 pKa = 11.01DD404 pKa = 3.64MGSAQALNTIDD415 pKa = 3.16QDD417 pKa = 3.94YY418 pKa = 10.72VRR420 pKa = 11.84RR421 pKa = 11.84AEE423 pKa = 4.12LATVAWGRR431 pKa = 11.84FQTQIADD438 pKa = 3.58LGITVGRR445 pKa = 11.84ALLPSLTDD453 pKa = 3.29LMNTVTPLIQRR464 pKa = 11.84TAQFAAAHH472 pKa = 6.54PGLIRR477 pKa = 11.84GVVGFATAVIGMKK490 pKa = 10.1VATLAAGWGLNFFVKK505 pKa = 10.55SPLNMVSTALTTVGAKK521 pKa = 6.89WTLFRR526 pKa = 11.84ALWAGGGSRR535 pKa = 11.84LSTVFQIFGMGAGAAGKK552 pKa = 8.95FAAVIGRR559 pKa = 11.84AGSLFMGFGRR569 pKa = 11.84GALVVGRR576 pKa = 11.84ALLPFGQGMLMTFIGPLRR594 pKa = 11.84LLAQGGMLLARR605 pKa = 11.84VLGGQLVNGLMLAGRR620 pKa = 11.84AVLWLGRR627 pKa = 11.84ALMLNPIGIAITAIAVGAYY646 pKa = 10.36LIYY649 pKa = 10.4RR650 pKa = 11.84YY651 pKa = 6.93WTPIKK656 pKa = 10.55QFFGGIWASIRR667 pKa = 11.84TAFAGVLGWFGVGLPKK683 pKa = 10.46TFTDD687 pKa = 4.81FGSHH691 pKa = 7.57LIDD694 pKa = 3.79GLVNGIRR701 pKa = 11.84NRR703 pKa = 11.84FTAAKK708 pKa = 7.81NTLIEE713 pKa = 4.47FGSNVKK719 pKa = 9.94AWFANTLGIKK729 pKa = 9.99SPSRR733 pKa = 11.84VFMGFGDD740 pKa = 4.89NIAQGAAIGIGRR752 pKa = 11.84SSAVAARR759 pKa = 11.84AAAGMATQAAAAASLQRR776 pKa = 11.84INAARR781 pKa = 11.84GGSPAGASVAGSGITVHH798 pKa = 6.72FSPTITVQGGSPDD811 pKa = 3.55GVKK814 pKa = 10.7DD815 pKa = 3.82QVKK818 pKa = 10.38QGLNLSLRR826 pKa = 11.84DD827 pKa = 4.07LEE829 pKa = 5.47RR830 pKa = 11.84MLDD833 pKa = 3.55DD834 pKa = 5.13LLAKK838 pKa = 10.08RR839 pKa = 11.84EE840 pKa = 3.81RR841 pKa = 11.84RR842 pKa = 11.84AYY844 pKa = 10.42RR845 pKa = 11.84SS846 pKa = 3.23
MM1 pKa = 7.57ASEE4 pKa = 4.93FYY6 pKa = 10.53IGVKK10 pKa = 10.01IGATLLGSFGAALSGTRR27 pKa = 11.84TTLNGLGRR35 pKa = 11.84VADD38 pKa = 4.21EE39 pKa = 3.62LRR41 pKa = 11.84ARR43 pKa = 11.84HH44 pKa = 5.45TRR46 pKa = 11.84LGDD49 pKa = 3.19AMARR53 pKa = 11.84AVAHH57 pKa = 6.07PMRR60 pKa = 11.84NIAEE64 pKa = 4.1LRR66 pKa = 11.84GQYY69 pKa = 10.5DD70 pKa = 3.37RR71 pKa = 11.84LGRR74 pKa = 11.84TIDD77 pKa = 3.44QVQAKK82 pKa = 8.79QAALATRR89 pKa = 11.84LARR92 pKa = 11.84GAALRR97 pKa = 11.84EE98 pKa = 3.92QRR100 pKa = 11.84QGLGADD106 pKa = 3.42MLGTYY111 pKa = 8.0ATAAATAAPVIGAVRR126 pKa = 11.84QAATFEE132 pKa = 4.2AGLRR136 pKa = 11.84DD137 pKa = 3.5IAITGNLTRR146 pKa = 11.84DD147 pKa = 3.09EE148 pKa = 4.22EE149 pKa = 4.22FRR151 pKa = 11.84IGEE154 pKa = 4.27TMRR157 pKa = 11.84RR158 pKa = 11.84AALATSQGHH167 pKa = 5.08NSILEE172 pKa = 4.22GVGTLVAAGMDD183 pKa = 3.48AKK185 pKa = 10.87EE186 pKa = 4.62AGQKK190 pKa = 10.64SNLLGRR196 pKa = 11.84VATATNADD204 pKa = 3.77MKK206 pKa = 10.84DD207 pKa = 3.54LAGMVYY213 pKa = 10.26SFSEE217 pKa = 4.06TLGIKK222 pKa = 10.17GDD224 pKa = 3.84AALKK228 pKa = 9.69EE229 pKa = 4.3AFNRR233 pKa = 11.84AAYY236 pKa = 9.71GGKK239 pKa = 10.07LGRR242 pKa = 11.84FEE244 pKa = 5.05LKK246 pKa = 10.8DD247 pKa = 3.31MAKK250 pKa = 10.47ALPEE254 pKa = 3.77MTAAFAAKK262 pKa = 10.08GIKK265 pKa = 10.08GQDD268 pKa = 3.2ALTQIIASLEE278 pKa = 4.07VGRR281 pKa = 11.84EE282 pKa = 3.94GAGSGDD288 pKa = 3.37EE289 pKa = 4.43AVTNLRR295 pKa = 11.84NWLSHH300 pKa = 5.73MNAKK304 pKa = 8.64ATIDD308 pKa = 3.52AYY310 pKa = 10.11KK311 pKa = 10.38KK312 pKa = 10.77AGVDD316 pKa = 3.73YY317 pKa = 10.29QKK319 pKa = 11.73SMSNLVAGGYY329 pKa = 9.46SSYY332 pKa = 11.01EE333 pKa = 3.64GSLQIAQKK341 pKa = 10.33FIASRR346 pKa = 11.84GDD348 pKa = 3.14AFMKK352 pKa = 9.23QWKK355 pKa = 9.13AAGAKK360 pKa = 9.93GDD362 pKa = 3.88EE363 pKa = 4.14EE364 pKa = 4.25AQRR367 pKa = 11.84KK368 pKa = 8.89LMEE371 pKa = 4.45SFGLNEE377 pKa = 4.17VFQDD381 pKa = 3.12IQTINHH387 pKa = 7.1LLAMRR392 pKa = 11.84QGWDD396 pKa = 3.12KK397 pKa = 11.1YY398 pKa = 7.82QQNKK402 pKa = 9.53KK403 pKa = 11.01DD404 pKa = 3.64MGSAQALNTIDD415 pKa = 3.16QDD417 pKa = 3.94YY418 pKa = 10.72VRR420 pKa = 11.84RR421 pKa = 11.84AEE423 pKa = 4.12LATVAWGRR431 pKa = 11.84FQTQIADD438 pKa = 3.58LGITVGRR445 pKa = 11.84ALLPSLTDD453 pKa = 3.29LMNTVTPLIQRR464 pKa = 11.84TAQFAAAHH472 pKa = 6.54PGLIRR477 pKa = 11.84GVVGFATAVIGMKK490 pKa = 10.1VATLAAGWGLNFFVKK505 pKa = 10.55SPLNMVSTALTTVGAKK521 pKa = 6.89WTLFRR526 pKa = 11.84ALWAGGGSRR535 pKa = 11.84LSTVFQIFGMGAGAAGKK552 pKa = 8.95FAAVIGRR559 pKa = 11.84AGSLFMGFGRR569 pKa = 11.84GALVVGRR576 pKa = 11.84ALLPFGQGMLMTFIGPLRR594 pKa = 11.84LLAQGGMLLARR605 pKa = 11.84VLGGQLVNGLMLAGRR620 pKa = 11.84AVLWLGRR627 pKa = 11.84ALMLNPIGIAITAIAVGAYY646 pKa = 10.36LIYY649 pKa = 10.4RR650 pKa = 11.84YY651 pKa = 6.93WTPIKK656 pKa = 10.55QFFGGIWASIRR667 pKa = 11.84TAFAGVLGWFGVGLPKK683 pKa = 10.46TFTDD687 pKa = 4.81FGSHH691 pKa = 7.57LIDD694 pKa = 3.79GLVNGIRR701 pKa = 11.84NRR703 pKa = 11.84FTAAKK708 pKa = 7.81NTLIEE713 pKa = 4.47FGSNVKK719 pKa = 9.94AWFANTLGIKK729 pKa = 9.99SPSRR733 pKa = 11.84VFMGFGDD740 pKa = 4.89NIAQGAAIGIGRR752 pKa = 11.84SSAVAARR759 pKa = 11.84AAAGMATQAAAAASLQRR776 pKa = 11.84INAARR781 pKa = 11.84GGSPAGASVAGSGITVHH798 pKa = 6.72FSPTITVQGGSPDD811 pKa = 3.55GVKK814 pKa = 10.7DD815 pKa = 3.82QVKK818 pKa = 10.38QGLNLSLRR826 pKa = 11.84DD827 pKa = 4.07LEE829 pKa = 5.47RR830 pKa = 11.84MLDD833 pKa = 3.55DD834 pKa = 5.13LLAKK838 pKa = 10.08RR839 pKa = 11.84EE840 pKa = 3.81RR841 pKa = 11.84RR842 pKa = 11.84AYY844 pKa = 10.42RR845 pKa = 11.84SS846 pKa = 3.23
Molecular weight: 89.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
11923 |
67 |
846 |
225.0 |
24.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.715 ± 0.468 | 0.646 ± 0.087 |
6.014 ± 0.326 | 5.661 ± 0.374 |
3.229 ± 0.22 | 8.11 ± 0.461 |
1.61 ± 0.177 | 4.697 ± 0.22 |
4.361 ± 0.307 | 9.134 ± 0.311 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.29 ± 0.165 | 3.095 ± 0.251 |
4.378 ± 0.26 | 4.202 ± 0.182 |
7.087 ± 0.39 | 5.603 ± 0.309 |
5.871 ± 0.399 | 7.314 ± 0.299 |
1.283 ± 0.131 | 2.701 ± 0.192 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
