
Termite associated circular virus 3
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
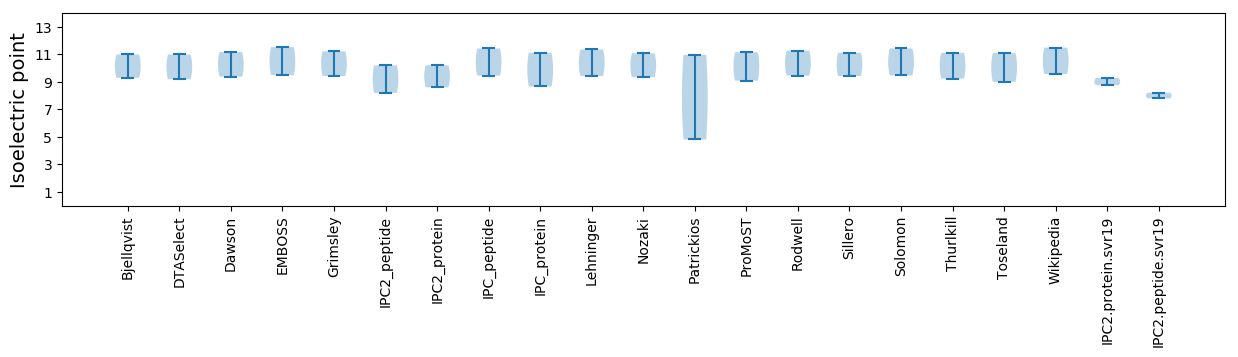
Average proteome isoelectric point is 9.02
Get precalculated fractions of proteins
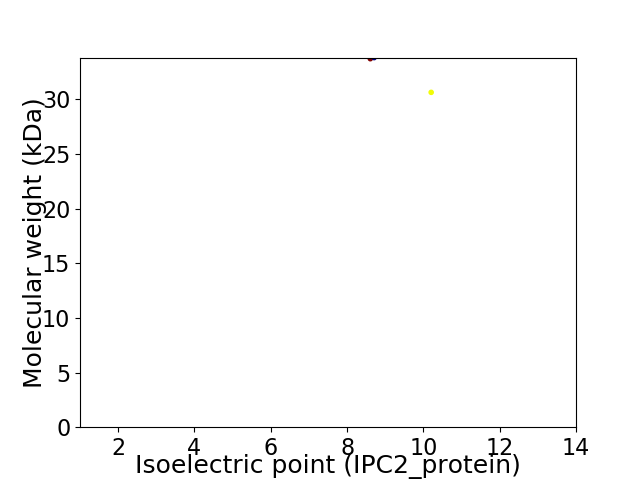
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2P1E318|A0A2P1E318_9VIRU Putative capsid protein OS=Termite associated circular virus 3 OX=2108551 PE=4 SV=1
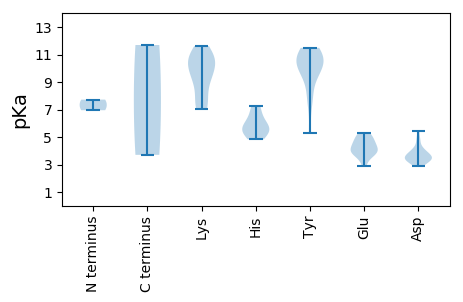
MM1 pKa = 7.72PFRR4 pKa = 11.84WQGKK8 pKa = 7.07YY9 pKa = 10.68LFLTYY14 pKa = 9.87PRR16 pKa = 11.84SDD18 pKa = 3.38FSLDD22 pKa = 3.16EE23 pKa = 4.17SLHH26 pKa = 5.44FFRR29 pKa = 11.84SQAKK33 pKa = 7.85QATFIRR39 pKa = 11.84VSSEE43 pKa = 3.39RR44 pKa = 11.84HH45 pKa = 5.41EE46 pKa = 5.3DD47 pKa = 3.71GSPHH51 pKa = 5.63RR52 pKa = 11.84HH53 pKa = 4.84VFIAFDD59 pKa = 3.38KK60 pKa = 10.82RR61 pKa = 11.84FSFTNEE67 pKa = 2.9RR68 pKa = 11.84RR69 pKa = 11.84FDD71 pKa = 3.46FRR73 pKa = 11.84EE74 pKa = 3.67RR75 pKa = 11.84HH76 pKa = 5.91PNIQSRR82 pKa = 11.84VDD84 pKa = 3.58NPAACLDD91 pKa = 3.98YY92 pKa = 11.03VSKK95 pKa = 11.25DD96 pKa = 3.46GVFVDD101 pKa = 4.27YY102 pKa = 11.4GSVPDD107 pKa = 4.88FNTSTSEE114 pKa = 3.75PRR116 pKa = 11.84EE117 pKa = 3.9SRR119 pKa = 11.84NQLWGRR125 pKa = 11.84LLDD128 pKa = 3.94EE129 pKa = 4.81ATSASHH135 pKa = 7.28FLQLVRR141 pKa = 11.84EE142 pKa = 3.99NSPYY146 pKa = 11.33DD147 pKa = 3.11FATRR151 pKa = 11.84VSSLFTIRR159 pKa = 11.84TAPYY163 pKa = 8.66AQNASSSSATLVPEE177 pKa = 4.63RR178 pKa = 11.84PAGRR182 pKa = 11.84GRR184 pKa = 11.84SGTISTSRR192 pKa = 11.84GSSTWTSLTPRR203 pKa = 11.84PATSSSTTARR213 pKa = 11.84SSGSRR218 pKa = 11.84TTSPGSGAGGEE229 pKa = 4.44FEE231 pKa = 5.02ATDD234 pKa = 3.48KK235 pKa = 11.38YY236 pKa = 10.48RR237 pKa = 11.84GKK239 pKa = 9.33RR240 pKa = 11.84TLVWQRR246 pKa = 11.84KK247 pKa = 7.06SCIVLCNRR255 pKa = 11.84GLLWDD260 pKa = 3.62WRR262 pKa = 11.84ASEE265 pKa = 4.9EE266 pKa = 4.39YY267 pKa = 10.31RR268 pKa = 11.84DD269 pKa = 3.75DD270 pKa = 5.43KK271 pKa = 11.61DD272 pKa = 3.55WFDD275 pKa = 4.7HH276 pKa = 6.51NVIICDD282 pKa = 3.95LNKK285 pKa = 10.51KK286 pKa = 8.34LWRR289 pKa = 11.84YY290 pKa = 9.93DD291 pKa = 3.12EE292 pKa = 5.26SYY294 pKa = 11.69
MM1 pKa = 7.72PFRR4 pKa = 11.84WQGKK8 pKa = 7.07YY9 pKa = 10.68LFLTYY14 pKa = 9.87PRR16 pKa = 11.84SDD18 pKa = 3.38FSLDD22 pKa = 3.16EE23 pKa = 4.17SLHH26 pKa = 5.44FFRR29 pKa = 11.84SQAKK33 pKa = 7.85QATFIRR39 pKa = 11.84VSSEE43 pKa = 3.39RR44 pKa = 11.84HH45 pKa = 5.41EE46 pKa = 5.3DD47 pKa = 3.71GSPHH51 pKa = 5.63RR52 pKa = 11.84HH53 pKa = 4.84VFIAFDD59 pKa = 3.38KK60 pKa = 10.82RR61 pKa = 11.84FSFTNEE67 pKa = 2.9RR68 pKa = 11.84RR69 pKa = 11.84FDD71 pKa = 3.46FRR73 pKa = 11.84EE74 pKa = 3.67RR75 pKa = 11.84HH76 pKa = 5.91PNIQSRR82 pKa = 11.84VDD84 pKa = 3.58NPAACLDD91 pKa = 3.98YY92 pKa = 11.03VSKK95 pKa = 11.25DD96 pKa = 3.46GVFVDD101 pKa = 4.27YY102 pKa = 11.4GSVPDD107 pKa = 4.88FNTSTSEE114 pKa = 3.75PRR116 pKa = 11.84EE117 pKa = 3.9SRR119 pKa = 11.84NQLWGRR125 pKa = 11.84LLDD128 pKa = 3.94EE129 pKa = 4.81ATSASHH135 pKa = 7.28FLQLVRR141 pKa = 11.84EE142 pKa = 3.99NSPYY146 pKa = 11.33DD147 pKa = 3.11FATRR151 pKa = 11.84VSSLFTIRR159 pKa = 11.84TAPYY163 pKa = 8.66AQNASSSSATLVPEE177 pKa = 4.63RR178 pKa = 11.84PAGRR182 pKa = 11.84GRR184 pKa = 11.84SGTISTSRR192 pKa = 11.84GSSTWTSLTPRR203 pKa = 11.84PATSSSTTARR213 pKa = 11.84SSGSRR218 pKa = 11.84TTSPGSGAGGEE229 pKa = 4.44FEE231 pKa = 5.02ATDD234 pKa = 3.48KK235 pKa = 11.38YY236 pKa = 10.48RR237 pKa = 11.84GKK239 pKa = 9.33RR240 pKa = 11.84TLVWQRR246 pKa = 11.84KK247 pKa = 7.06SCIVLCNRR255 pKa = 11.84GLLWDD260 pKa = 3.62WRR262 pKa = 11.84ASEE265 pKa = 4.9EE266 pKa = 4.39YY267 pKa = 10.31RR268 pKa = 11.84DD269 pKa = 3.75DD270 pKa = 5.43KK271 pKa = 11.61DD272 pKa = 3.55WFDD275 pKa = 4.7HH276 pKa = 6.51NVIICDD282 pKa = 3.95LNKK285 pKa = 10.51KK286 pKa = 8.34LWRR289 pKa = 11.84YY290 pKa = 9.93DD291 pKa = 3.12EE292 pKa = 5.26SYY294 pKa = 11.69
Molecular weight: 33.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2P1E318|A0A2P1E318_9VIRU Putative capsid protein OS=Termite associated circular virus 3 OX=2108551 PE=4 SV=1
MM1 pKa = 6.95PQRR4 pKa = 11.84YY5 pKa = 5.29TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84YY12 pKa = 7.54RR13 pKa = 11.84STRR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84KK20 pKa = 7.57VVRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VPKK29 pKa = 9.5ATMKK33 pKa = 9.8MVRR36 pKa = 11.84SVANRR41 pKa = 11.84QIMKK45 pKa = 9.27ATEE48 pKa = 3.86TKK50 pKa = 10.28RR51 pKa = 11.84NVRR54 pKa = 11.84LEE56 pKa = 4.48EE57 pKa = 3.91NWSPIPSNSTGSQYY71 pKa = 10.97TIANIFAQIGGGGTASGTNATQLIGDD97 pKa = 4.15TFFNPLFKK105 pKa = 10.93AKK107 pKa = 10.02LRR109 pKa = 11.84YY110 pKa = 9.45FIDD113 pKa = 3.38WNRR116 pKa = 11.84VRR118 pKa = 11.84SLNGSGTGVLPVQLYY133 pKa = 9.57CWIVAANDD141 pKa = 3.56ILPLGVTPNNYY152 pKa = 7.81FTISPQPNFFLTSGGTAPTFNGDD175 pKa = 3.01NVRR178 pKa = 11.84VIRR181 pKa = 11.84KK182 pKa = 7.04WSKK185 pKa = 9.37TINPPSIVVGAGTVVTNGASLYY207 pKa = 10.38KK208 pKa = 10.62KK209 pKa = 10.34NITVKK214 pKa = 10.3FRR216 pKa = 11.84GSKK219 pKa = 8.64TFEE222 pKa = 4.15SDD224 pKa = 2.91SSGGLGFGLRR234 pKa = 11.84GWNFYY239 pKa = 10.26MITGYY244 pKa = 11.49AMPYY248 pKa = 10.19GFALSGTPLLPLVDD262 pKa = 3.89VRR264 pKa = 11.84VDD266 pKa = 3.28RR267 pKa = 11.84YY268 pKa = 11.1LYY270 pKa = 10.79FKK272 pKa = 10.97DD273 pKa = 3.47PP274 pKa = 3.7
MM1 pKa = 6.95PQRR4 pKa = 11.84YY5 pKa = 5.29TRR7 pKa = 11.84RR8 pKa = 11.84RR9 pKa = 11.84SRR11 pKa = 11.84YY12 pKa = 7.54RR13 pKa = 11.84STRR16 pKa = 11.84RR17 pKa = 11.84VRR19 pKa = 11.84KK20 pKa = 7.57VVRR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84VPKK29 pKa = 9.5ATMKK33 pKa = 9.8MVRR36 pKa = 11.84SVANRR41 pKa = 11.84QIMKK45 pKa = 9.27ATEE48 pKa = 3.86TKK50 pKa = 10.28RR51 pKa = 11.84NVRR54 pKa = 11.84LEE56 pKa = 4.48EE57 pKa = 3.91NWSPIPSNSTGSQYY71 pKa = 10.97TIANIFAQIGGGGTASGTNATQLIGDD97 pKa = 4.15TFFNPLFKK105 pKa = 10.93AKK107 pKa = 10.02LRR109 pKa = 11.84YY110 pKa = 9.45FIDD113 pKa = 3.38WNRR116 pKa = 11.84VRR118 pKa = 11.84SLNGSGTGVLPVQLYY133 pKa = 9.57CWIVAANDD141 pKa = 3.56ILPLGVTPNNYY152 pKa = 7.81FTISPQPNFFLTSGGTAPTFNGDD175 pKa = 3.01NVRR178 pKa = 11.84VIRR181 pKa = 11.84KK182 pKa = 7.04WSKK185 pKa = 9.37TINPPSIVVGAGTVVTNGASLYY207 pKa = 10.38KK208 pKa = 10.62KK209 pKa = 10.34NITVKK214 pKa = 10.3FRR216 pKa = 11.84GSKK219 pKa = 8.64TFEE222 pKa = 4.15SDD224 pKa = 2.91SSGGLGFGLRR234 pKa = 11.84GWNFYY239 pKa = 10.26MITGYY244 pKa = 11.49AMPYY248 pKa = 10.19GFALSGTPLLPLVDD262 pKa = 3.89VRR264 pKa = 11.84VDD266 pKa = 3.28RR267 pKa = 11.84YY268 pKa = 11.1LYY270 pKa = 10.79FKK272 pKa = 10.97DD273 pKa = 3.47PP274 pKa = 3.7
Molecular weight: 30.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
568 |
274 |
294 |
284.0 |
32.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.81 ± 0.215 | 0.88 ± 0.33 |
4.93 ± 1.288 | 3.345 ± 1.208 |
5.986 ± 0.328 | 7.394 ± 1.343 |
1.232 ± 0.79 | 4.049 ± 0.913 |
4.225 ± 0.567 | 6.514 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.232 ± 0.614 | 5.106 ± 1.172 |
5.458 ± 0.479 | 2.641 ± 0.055 |
10.211 ± 0.463 | 10.563 ± 2.092 |
8.099 ± 0.423 | 6.162 ± 1.197 |
2.289 ± 0.297 | 3.873 ± 0.325 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
