
Lynx canadensis faeces associated genomovirus CL3 128
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.35
Get precalculated fractions of proteins
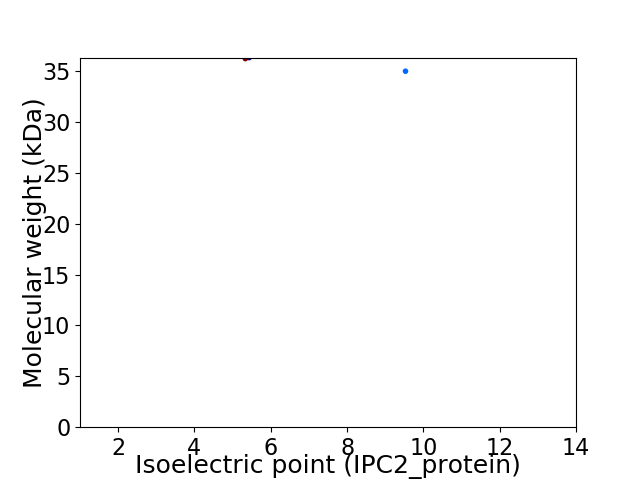
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2Z5CI23|A0A2Z5CI23_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL3 128 OX=2219129 PE=3 SV=1
MM1 pKa = 7.83APFFLKK7 pKa = 10.17NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.97VMLTYY16 pKa = 10.52SQAGSDD22 pKa = 3.46FDD24 pKa = 3.5YY25 pKa = 10.94WSIVEE30 pKa = 4.47LLSSHH35 pKa = 6.0GAEE38 pKa = 4.28CIIGRR43 pKa = 11.84EE44 pKa = 4.02LHH46 pKa = 6.86ADD48 pKa = 3.64GGTHH52 pKa = 5.47FHH54 pKa = 6.24VFVDD58 pKa = 4.64FGRR61 pKa = 11.84LFQRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.43TDD69 pKa = 3.35LFDD72 pKa = 4.17VGGKK76 pKa = 9.23HH77 pKa = 6.12PNIMPVWKK85 pKa = 8.88TPGEE89 pKa = 4.12AFDD92 pKa = 4.3YY93 pKa = 8.57ATKK96 pKa = 10.85DD97 pKa = 3.15GDD99 pKa = 3.7IVAGGLEE106 pKa = 4.42RR107 pKa = 11.84PGTDD111 pKa = 3.49CDD113 pKa = 4.09YY114 pKa = 10.36DD115 pKa = 3.91TTNFWATAGASQSGEE130 pKa = 4.12EE131 pKa = 4.25FLHH134 pKa = 7.05FLDD137 pKa = 4.3QLAPRR142 pKa = 11.84DD143 pKa = 3.75LMRR146 pKa = 11.84GFLQFRR152 pKa = 11.84SYY154 pKa = 11.73ADD156 pKa = 3.41WKK158 pKa = 9.37WAVAPEE164 pKa = 4.45RR165 pKa = 11.84YY166 pKa = 9.28VDD168 pKa = 3.61PPGVVFDD175 pKa = 4.33TGHH178 pKa = 7.08AEE180 pKa = 3.85QLSEE184 pKa = 4.59RR185 pKa = 11.84KK186 pKa = 9.83SLMLWGPSQFGKK198 pKa = 6.66TTWARR203 pKa = 11.84SLGNHH208 pKa = 6.2IFFGSQFSGKK218 pKa = 9.73LALDD222 pKa = 4.0GMDD225 pKa = 3.8DD226 pKa = 3.34AQYY229 pKa = 11.33AIFDD233 pKa = 3.77DD234 pKa = 3.97WKK236 pKa = 10.99GGMKK240 pKa = 10.47ALPGYY245 pKa = 9.93KK246 pKa = 9.62DD247 pKa = 2.93WFGCQWQISVRR258 pKa = 11.84KK259 pKa = 8.42LHH261 pKa = 6.73HH262 pKa = 7.08DD263 pKa = 3.06ARR265 pKa = 11.84LITWGRR271 pKa = 11.84PIIWLCNKK279 pKa = 9.89DD280 pKa = 3.61PRR282 pKa = 11.84LMHH285 pKa = 6.99IATDD289 pKa = 3.82DD290 pKa = 4.26VDD292 pKa = 4.07WEE294 pKa = 4.3WMDD297 pKa = 3.67EE298 pKa = 3.69NVMFVEE304 pKa = 4.79IVRR307 pKa = 11.84PLVTFHH313 pKa = 7.37ANTT316 pKa = 3.62
MM1 pKa = 7.83APFFLKK7 pKa = 10.17NRR9 pKa = 11.84RR10 pKa = 11.84YY11 pKa = 10.97VMLTYY16 pKa = 10.52SQAGSDD22 pKa = 3.46FDD24 pKa = 3.5YY25 pKa = 10.94WSIVEE30 pKa = 4.47LLSSHH35 pKa = 6.0GAEE38 pKa = 4.28CIIGRR43 pKa = 11.84EE44 pKa = 4.02LHH46 pKa = 6.86ADD48 pKa = 3.64GGTHH52 pKa = 5.47FHH54 pKa = 6.24VFVDD58 pKa = 4.64FGRR61 pKa = 11.84LFQRR65 pKa = 11.84RR66 pKa = 11.84KK67 pKa = 8.43TDD69 pKa = 3.35LFDD72 pKa = 4.17VGGKK76 pKa = 9.23HH77 pKa = 6.12PNIMPVWKK85 pKa = 8.88TPGEE89 pKa = 4.12AFDD92 pKa = 4.3YY93 pKa = 8.57ATKK96 pKa = 10.85DD97 pKa = 3.15GDD99 pKa = 3.7IVAGGLEE106 pKa = 4.42RR107 pKa = 11.84PGTDD111 pKa = 3.49CDD113 pKa = 4.09YY114 pKa = 10.36DD115 pKa = 3.91TTNFWATAGASQSGEE130 pKa = 4.12EE131 pKa = 4.25FLHH134 pKa = 7.05FLDD137 pKa = 4.3QLAPRR142 pKa = 11.84DD143 pKa = 3.75LMRR146 pKa = 11.84GFLQFRR152 pKa = 11.84SYY154 pKa = 11.73ADD156 pKa = 3.41WKK158 pKa = 9.37WAVAPEE164 pKa = 4.45RR165 pKa = 11.84YY166 pKa = 9.28VDD168 pKa = 3.61PPGVVFDD175 pKa = 4.33TGHH178 pKa = 7.08AEE180 pKa = 3.85QLSEE184 pKa = 4.59RR185 pKa = 11.84KK186 pKa = 9.83SLMLWGPSQFGKK198 pKa = 6.66TTWARR203 pKa = 11.84SLGNHH208 pKa = 6.2IFFGSQFSGKK218 pKa = 9.73LALDD222 pKa = 4.0GMDD225 pKa = 3.8DD226 pKa = 3.34AQYY229 pKa = 11.33AIFDD233 pKa = 3.77DD234 pKa = 3.97WKK236 pKa = 10.99GGMKK240 pKa = 10.47ALPGYY245 pKa = 9.93KK246 pKa = 9.62DD247 pKa = 2.93WFGCQWQISVRR258 pKa = 11.84KK259 pKa = 8.42LHH261 pKa = 6.73HH262 pKa = 7.08DD263 pKa = 3.06ARR265 pKa = 11.84LITWGRR271 pKa = 11.84PIIWLCNKK279 pKa = 9.89DD280 pKa = 3.61PRR282 pKa = 11.84LMHH285 pKa = 6.99IATDD289 pKa = 3.82DD290 pKa = 4.26VDD292 pKa = 4.07WEE294 pKa = 4.3WMDD297 pKa = 3.67EE298 pKa = 3.69NVMFVEE304 pKa = 4.79IVRR307 pKa = 11.84PLVTFHH313 pKa = 7.37ANTT316 pKa = 3.62
Molecular weight: 36.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CI23|A0A2Z5CI23_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL3 128 OX=2219129 PE=3 SV=1
MM1 pKa = 7.83AYY3 pKa = 9.73KK4 pKa = 10.34SYY6 pKa = 11.19AKK8 pKa = 10.04RR9 pKa = 11.84RR10 pKa = 11.84PTRR13 pKa = 11.84RR14 pKa = 11.84SYY16 pKa = 10.39KK17 pKa = 9.4SKK19 pKa = 10.33RR20 pKa = 11.84RR21 pKa = 11.84SAPRR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.87AKK30 pKa = 10.01KK31 pKa = 10.08RR32 pKa = 11.84STFPSRR38 pKa = 11.84RR39 pKa = 11.84MSKK42 pKa = 10.44KK43 pKa = 9.98KK44 pKa = 9.74ILNVVSRR51 pKa = 11.84KK52 pKa = 9.29KK53 pKa = 10.47RR54 pKa = 11.84NGMMSWSNTTSSSGASQSIAAGNAVVAGNSGGTFLFSPTCMNLNQGTTSPNYY106 pKa = 9.95AINVAEE112 pKa = 4.33RR113 pKa = 11.84TATTCYY119 pKa = 9.81MKK121 pKa = 10.7GFKK124 pKa = 10.32EE125 pKa = 4.11NIRR128 pKa = 11.84IQTNSHH134 pKa = 6.26LPWFHH139 pKa = 6.69RR140 pKa = 11.84RR141 pKa = 11.84ICFRR145 pKa = 11.84YY146 pKa = 9.17RR147 pKa = 11.84GSGPFTTANPVDD159 pKa = 4.11TPTNTVTPYY168 pKa = 10.26IDD170 pKa = 3.37TSNGMEE176 pKa = 4.19RR177 pKa = 11.84LWLNQQVNNMPQTLVVLNQILFKK200 pKa = 10.95GSQNQDD206 pKa = 2.32WNDD209 pKa = 3.13IVIAPVDD216 pKa = 3.6TARR219 pKa = 11.84VDD221 pKa = 4.02LVFDD225 pKa = 3.76KK226 pKa = 10.78TWLIKK231 pKa = 10.64SGNEE235 pKa = 3.29TGTILEE241 pKa = 4.16RR242 pKa = 11.84KK243 pKa = 8.6LWHH246 pKa = 6.39GMNKK250 pKa = 9.72NLVFDD255 pKa = 4.85DD256 pKa = 4.7DD257 pKa = 5.07EE258 pKa = 6.25IGEE261 pKa = 4.57SMNSNYY267 pKa = 9.84FSVDD271 pKa = 2.98SKK273 pKa = 11.36RR274 pKa = 11.84GMGDD278 pKa = 3.13FYY280 pKa = 10.57IYY282 pKa = 10.43DD283 pKa = 3.6IVQPGLGGGATDD295 pKa = 6.39LISLSANSTMYY306 pKa = 9.46WHH308 pKa = 7.07EE309 pKa = 4.05KK310 pKa = 9.5
MM1 pKa = 7.83AYY3 pKa = 9.73KK4 pKa = 10.34SYY6 pKa = 11.19AKK8 pKa = 10.04RR9 pKa = 11.84RR10 pKa = 11.84PTRR13 pKa = 11.84RR14 pKa = 11.84SYY16 pKa = 10.39KK17 pKa = 9.4SKK19 pKa = 10.33RR20 pKa = 11.84RR21 pKa = 11.84SAPRR25 pKa = 11.84RR26 pKa = 11.84SYY28 pKa = 10.87AKK30 pKa = 10.01KK31 pKa = 10.08RR32 pKa = 11.84STFPSRR38 pKa = 11.84RR39 pKa = 11.84MSKK42 pKa = 10.44KK43 pKa = 9.98KK44 pKa = 9.74ILNVVSRR51 pKa = 11.84KK52 pKa = 9.29KK53 pKa = 10.47RR54 pKa = 11.84NGMMSWSNTTSSSGASQSIAAGNAVVAGNSGGTFLFSPTCMNLNQGTTSPNYY106 pKa = 9.95AINVAEE112 pKa = 4.33RR113 pKa = 11.84TATTCYY119 pKa = 9.81MKK121 pKa = 10.7GFKK124 pKa = 10.32EE125 pKa = 4.11NIRR128 pKa = 11.84IQTNSHH134 pKa = 6.26LPWFHH139 pKa = 6.69RR140 pKa = 11.84RR141 pKa = 11.84ICFRR145 pKa = 11.84YY146 pKa = 9.17RR147 pKa = 11.84GSGPFTTANPVDD159 pKa = 4.11TPTNTVTPYY168 pKa = 10.26IDD170 pKa = 3.37TSNGMEE176 pKa = 4.19RR177 pKa = 11.84LWLNQQVNNMPQTLVVLNQILFKK200 pKa = 10.95GSQNQDD206 pKa = 2.32WNDD209 pKa = 3.13IVIAPVDD216 pKa = 3.6TARR219 pKa = 11.84VDD221 pKa = 4.02LVFDD225 pKa = 3.76KK226 pKa = 10.78TWLIKK231 pKa = 10.64SGNEE235 pKa = 3.29TGTILEE241 pKa = 4.16RR242 pKa = 11.84KK243 pKa = 8.6LWHH246 pKa = 6.39GMNKK250 pKa = 9.72NLVFDD255 pKa = 4.85DD256 pKa = 4.7DD257 pKa = 5.07EE258 pKa = 6.25IGEE261 pKa = 4.57SMNSNYY267 pKa = 9.84FSVDD271 pKa = 2.98SKK273 pKa = 11.36RR274 pKa = 11.84GMGDD278 pKa = 3.13FYY280 pKa = 10.57IYY282 pKa = 10.43DD283 pKa = 3.6IVQPGLGGGATDD295 pKa = 6.39LISLSANSTMYY306 pKa = 9.46WHH308 pKa = 7.07EE309 pKa = 4.05KK310 pKa = 9.5
Molecular weight: 35.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
626 |
310 |
316 |
313.0 |
35.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.39 ± 0.608 | 1.118 ± 0.101 |
6.869 ± 1.579 | 3.355 ± 0.519 |
5.591 ± 1.154 | 8.147 ± 0.705 |
2.556 ± 0.849 | 4.633 ± 0.355 |
5.431 ± 0.685 | 6.709 ± 0.822 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.514 ± 0.239 | 5.272 ± 2.09 |
4.313 ± 0.08 | 3.355 ± 0.086 |
6.55 ± 0.584 | 7.348 ± 1.779 |
6.709 ± 1.126 | 5.431 ± 0.035 |
3.355 ± 0.736 | 3.355 ± 0.346 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
