
Phytophthora ramorum (Sudden oak death agent)
Taxonomy: cellular organisms; Eukaryota; Sar; Stramenopiles; Oomycota; Peronosporales; Peronosporaceae; Phytophthora
Average proteome isoelectric point is 6.3
Get precalculated fractions of proteins
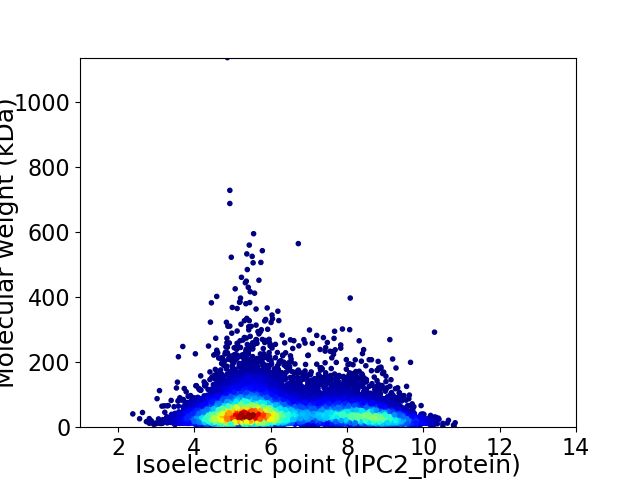
Virtual 2D-PAGE plot for 15349 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|H3G7L4|H3G7L4_PHYRM Nucleolar GTP-binding protein 2 OS=Phytophthora ramorum OX=164328 PE=3 SV=1
TT1 pKa = 7.09AACTLSGTYY10 pKa = 10.22SDD12 pKa = 4.03GTDD15 pKa = 3.03VSSCSSIVISDD26 pKa = 3.36LTVPKK31 pKa = 10.63GVTLDD36 pKa = 3.67LTNVADD42 pKa = 4.17GATITFEE49 pKa = 3.92GTTTFGEE56 pKa = 4.48EE57 pKa = 4.07EE58 pKa = 3.96WDD60 pKa = 3.87GPLILLTGNDD70 pKa = 3.62LTVTGTGTLDD80 pKa = 4.34GQGSWYY86 pKa = 9.14WEE88 pKa = 3.8QGTSISKK95 pKa = 9.97PVFFRR100 pKa = 11.84LKK102 pKa = 10.44KK103 pKa = 10.33VMSSTLEE110 pKa = 3.79SFNIKK115 pKa = 9.94NSPYY119 pKa = 8.82RR120 pKa = 11.84TFSILDD126 pKa = 3.85CEE128 pKa = 4.46DD129 pKa = 3.31TTLTGLTLDD138 pKa = 4.35SSAGDD143 pKa = 3.54DD144 pKa = 3.58TAKK147 pKa = 9.69NTDD150 pKa = 3.26GFDD153 pKa = 3.3LSRR156 pKa = 11.84NVGLTISDD164 pKa = 3.69CTVYY168 pKa = 11.07NQDD171 pKa = 4.36DD172 pKa = 4.37CLAMQSSNDD181 pKa = 3.46TTFSGNTCSGGHH193 pKa = 6.71GISIGSLGGDD203 pKa = 3.09SVTEE207 pKa = 3.85SDD209 pKa = 3.54IVSGLTVKK217 pKa = 10.44NNKK220 pKa = 9.82IIDD223 pKa = 3.99SVNGIRR229 pKa = 11.84IKK231 pKa = 10.11TIIDD235 pKa = 3.96LYY237 pKa = 10.07GTVTGASYY245 pKa = 11.19TDD247 pKa = 3.31NTLSNVEE254 pKa = 3.94NAIVIHH260 pKa = 6.36SDD262 pKa = 3.35YY263 pKa = 11.08SKK265 pKa = 11.39SEE267 pKa = 3.57GGYY270 pKa = 8.8TGDD273 pKa = 3.59ATSKK277 pKa = 10.72VAITDD282 pKa = 3.23ITISGLSGTADD293 pKa = 4.47DD294 pKa = 5.47IYY296 pKa = 11.12DD297 pKa = 3.55ILVNADD303 pKa = 3.75VVSGWTFSDD312 pKa = 3.12ITVTGDD318 pKa = 2.87TGSCSGEE325 pKa = 3.86PSGVDD330 pKa = 3.26CC331 pKa = 6.25
TT1 pKa = 7.09AACTLSGTYY10 pKa = 10.22SDD12 pKa = 4.03GTDD15 pKa = 3.03VSSCSSIVISDD26 pKa = 3.36LTVPKK31 pKa = 10.63GVTLDD36 pKa = 3.67LTNVADD42 pKa = 4.17GATITFEE49 pKa = 3.92GTTTFGEE56 pKa = 4.48EE57 pKa = 4.07EE58 pKa = 3.96WDD60 pKa = 3.87GPLILLTGNDD70 pKa = 3.62LTVTGTGTLDD80 pKa = 4.34GQGSWYY86 pKa = 9.14WEE88 pKa = 3.8QGTSISKK95 pKa = 9.97PVFFRR100 pKa = 11.84LKK102 pKa = 10.44KK103 pKa = 10.33VMSSTLEE110 pKa = 3.79SFNIKK115 pKa = 9.94NSPYY119 pKa = 8.82RR120 pKa = 11.84TFSILDD126 pKa = 3.85CEE128 pKa = 4.46DD129 pKa = 3.31TTLTGLTLDD138 pKa = 4.35SSAGDD143 pKa = 3.54DD144 pKa = 3.58TAKK147 pKa = 9.69NTDD150 pKa = 3.26GFDD153 pKa = 3.3LSRR156 pKa = 11.84NVGLTISDD164 pKa = 3.69CTVYY168 pKa = 11.07NQDD171 pKa = 4.36DD172 pKa = 4.37CLAMQSSNDD181 pKa = 3.46TTFSGNTCSGGHH193 pKa = 6.71GISIGSLGGDD203 pKa = 3.09SVTEE207 pKa = 3.85SDD209 pKa = 3.54IVSGLTVKK217 pKa = 10.44NNKK220 pKa = 9.82IIDD223 pKa = 3.99SVNGIRR229 pKa = 11.84IKK231 pKa = 10.11TIIDD235 pKa = 3.96LYY237 pKa = 10.07GTVTGASYY245 pKa = 11.19TDD247 pKa = 3.31NTLSNVEE254 pKa = 3.94NAIVIHH260 pKa = 6.36SDD262 pKa = 3.35YY263 pKa = 11.08SKK265 pKa = 11.39SEE267 pKa = 3.57GGYY270 pKa = 8.8TGDD273 pKa = 3.59ATSKK277 pKa = 10.72VAITDD282 pKa = 3.23ITISGLSGTADD293 pKa = 4.47DD294 pKa = 5.47IYY296 pKa = 11.12DD297 pKa = 3.55ILVNADD303 pKa = 3.75VVSGWTFSDD312 pKa = 3.12ITVTGDD318 pKa = 2.87TGSCSGEE325 pKa = 3.86PSGVDD330 pKa = 3.26CC331 pKa = 6.25
Molecular weight: 34.44 kDa
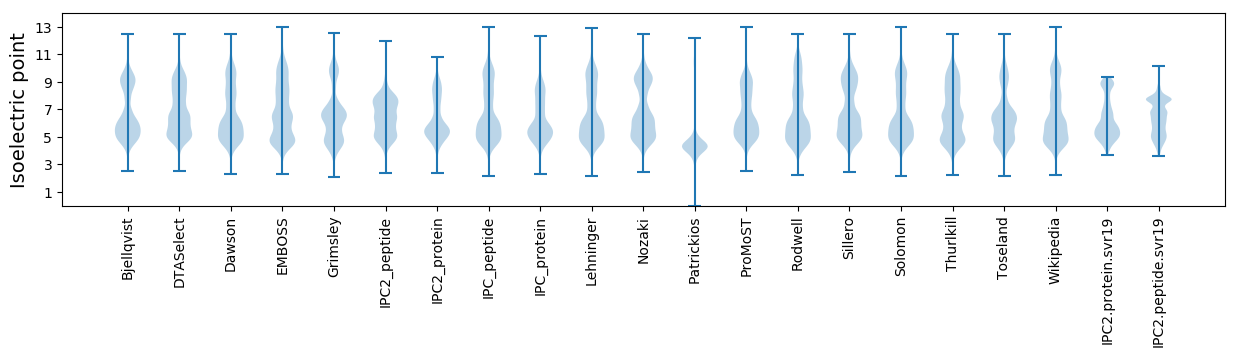
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|H3G5I3|H3G5I3_PHYRM 40S ribosomal protein S26 OS=Phytophthora ramorum OX=164328 PE=3 SV=1
MM1 pKa = 7.36VKK3 pKa = 10.16QKK5 pKa = 10.61NHH7 pKa = 4.55TARR10 pKa = 11.84NNTVKK15 pKa = 10.67AHH17 pKa = 6.51RR18 pKa = 11.84NGIKK22 pKa = 9.98KK23 pKa = 9.79PKK25 pKa = 8.2NHH27 pKa = 6.98RR28 pKa = 11.84FHH30 pKa = 6.43STRR33 pKa = 11.84GVV35 pKa = 2.99
MM1 pKa = 7.36VKK3 pKa = 10.16QKK5 pKa = 10.61NHH7 pKa = 4.55TARR10 pKa = 11.84NNTVKK15 pKa = 10.67AHH17 pKa = 6.51RR18 pKa = 11.84NGIKK22 pKa = 9.98KK23 pKa = 9.79PKK25 pKa = 8.2NHH27 pKa = 6.98RR28 pKa = 11.84FHH30 pKa = 6.43STRR33 pKa = 11.84GVV35 pKa = 2.99
Molecular weight: 4.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
7381725 |
34 |
10810 |
480.9 |
53.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.424 ± 0.02 | 1.56 ± 0.009 |
5.828 ± 0.013 | 6.695 ± 0.023 |
3.734 ± 0.014 | 6.369 ± 0.021 |
2.292 ± 0.008 | 3.83 ± 0.013 |
5.029 ± 0.017 | 9.27 ± 0.019 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.009 | 3.335 ± 0.009 |
4.75 ± 0.017 | 4.2 ± 0.017 |
6.122 ± 0.018 | 8.089 ± 0.029 |
5.924 ± 0.019 | 7.25 ± 0.016 |
1.222 ± 0.006 | 2.61 ± 0.011 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
