
Ranid herpesvirus 1 (strain McKinnell) (RaHV-1)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Alloherpesviridae; Batrachovirus; Ranid herpesvirus 1
Average proteome isoelectric point is 7.18
Get precalculated fractions of proteins
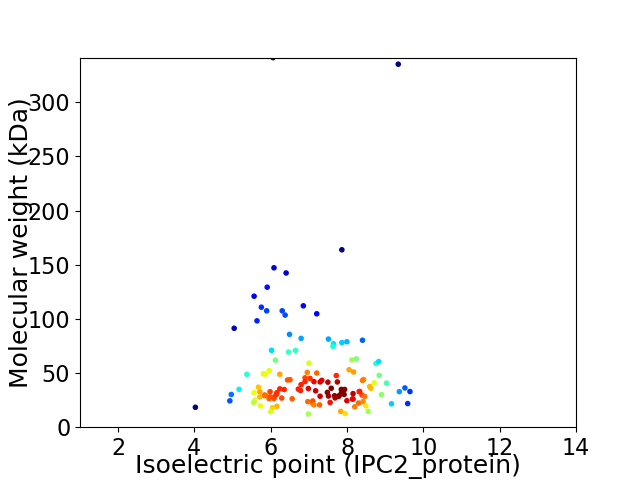
Virtual 2D-PAGE plot for 132 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|Q9YQZ9|Q9YQZ9_RAHV1 ORF69 OS=Ranid herpesvirus 1 (strain McKinnell) OX=1283342 PE=4 SV=1
MM1 pKa = 7.49EE2 pKa = 5.64SSLEE6 pKa = 3.97MSSEE10 pKa = 3.96DD11 pKa = 4.0QEE13 pKa = 4.87NIAAGHH19 pKa = 6.88EE20 pKa = 3.99IQIIADD26 pKa = 3.48AFVSNGYY33 pKa = 10.26LSRR36 pKa = 11.84TQAFMATCVGMLMASIGQEE55 pKa = 3.89FEE57 pKa = 3.7AAEE60 pKa = 4.24PRR62 pKa = 11.84DD63 pKa = 3.86VDD65 pKa = 4.11YY66 pKa = 11.01MFDD69 pKa = 3.73DD70 pKa = 4.06RR71 pKa = 11.84TDD73 pKa = 3.37EE74 pKa = 4.8AVDD77 pKa = 3.56HH78 pKa = 6.11MLHH81 pKa = 7.07LFLDD85 pKa = 4.8DD86 pKa = 4.3AAHH89 pKa = 7.27AYY91 pKa = 8.89AKK93 pKa = 10.78CEE95 pKa = 3.96LDD97 pKa = 3.33EE98 pKa = 4.54FLAGSAEE105 pKa = 4.04EE106 pKa = 4.16QEE108 pKa = 4.47RR109 pKa = 11.84EE110 pKa = 3.95LAEE113 pKa = 4.29EE114 pKa = 4.58GSVIVEE120 pKa = 4.81DD121 pKa = 4.3YY122 pKa = 10.95PLTDD126 pKa = 3.09VTGFAVFTPSSDD138 pKa = 3.29APIVIVGVCKK148 pKa = 9.84DD149 pKa = 3.63TSWRR153 pKa = 11.84HH154 pKa = 4.82ADD156 pKa = 3.05IANLVHH162 pKa = 6.65RR163 pKa = 11.84VLMSS167 pKa = 3.57
MM1 pKa = 7.49EE2 pKa = 5.64SSLEE6 pKa = 3.97MSSEE10 pKa = 3.96DD11 pKa = 4.0QEE13 pKa = 4.87NIAAGHH19 pKa = 6.88EE20 pKa = 3.99IQIIADD26 pKa = 3.48AFVSNGYY33 pKa = 10.26LSRR36 pKa = 11.84TQAFMATCVGMLMASIGQEE55 pKa = 3.89FEE57 pKa = 3.7AAEE60 pKa = 4.24PRR62 pKa = 11.84DD63 pKa = 3.86VDD65 pKa = 4.11YY66 pKa = 11.01MFDD69 pKa = 3.73DD70 pKa = 4.06RR71 pKa = 11.84TDD73 pKa = 3.37EE74 pKa = 4.8AVDD77 pKa = 3.56HH78 pKa = 6.11MLHH81 pKa = 7.07LFLDD85 pKa = 4.8DD86 pKa = 4.3AAHH89 pKa = 7.27AYY91 pKa = 8.89AKK93 pKa = 10.78CEE95 pKa = 3.96LDD97 pKa = 3.33EE98 pKa = 4.54FLAGSAEE105 pKa = 4.04EE106 pKa = 4.16QEE108 pKa = 4.47RR109 pKa = 11.84EE110 pKa = 3.95LAEE113 pKa = 4.29EE114 pKa = 4.58GSVIVEE120 pKa = 4.81DD121 pKa = 4.3YY122 pKa = 10.95PLTDD126 pKa = 3.09VTGFAVFTPSSDD138 pKa = 3.29APIVIVGVCKK148 pKa = 9.84DD149 pKa = 3.63TSWRR153 pKa = 11.84HH154 pKa = 4.82ADD156 pKa = 3.05IANLVHH162 pKa = 6.65RR163 pKa = 11.84VLMSS167 pKa = 3.57
Molecular weight: 18.4 kDa
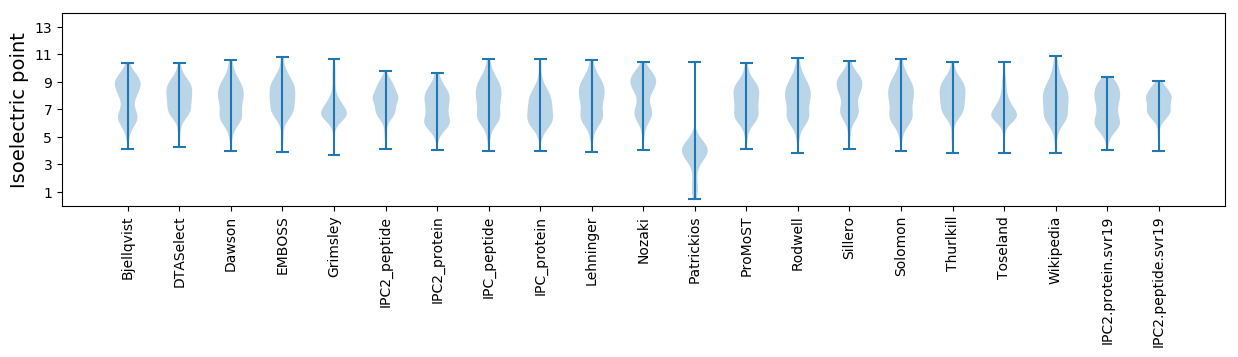
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q14VR3|Q14VR3_RAHV1 ORF45 OS=Ranid herpesvirus 1 (strain McKinnell) OX=1283342 PE=4 SV=1
MM1 pKa = 7.32SRR3 pKa = 11.84LMLTGVRR10 pKa = 11.84PSASPVSMHH19 pKa = 6.32TPAAHH24 pKa = 6.69CAPSSSAVVHH34 pKa = 5.82LHH36 pKa = 5.79SAAIHH41 pKa = 4.97MAKK44 pKa = 10.16AVSTCPGARR53 pKa = 11.84SKK55 pKa = 10.94AARR58 pKa = 11.84NSSLIGSTNRR68 pKa = 11.84LPTQPITSAHH78 pKa = 6.35IPRR81 pKa = 11.84TSGDD85 pKa = 3.14TSSPPNSSTILATYY99 pKa = 9.11KK100 pKa = 10.65ARR102 pKa = 11.84AISSNDD108 pKa = 2.51IPRR111 pKa = 11.84FYY113 pKa = 10.92GGRR116 pKa = 11.84YY117 pKa = 6.2TRR119 pKa = 11.84ARR121 pKa = 11.84EE122 pKa = 3.46QAQYY126 pKa = 10.65AKK128 pKa = 9.9HH129 pKa = 5.37GVGMPRR135 pKa = 11.84RR136 pKa = 11.84LRR138 pKa = 11.84AALWDD143 pKa = 3.66RR144 pKa = 11.84TVCSAHH150 pKa = 6.95RR151 pKa = 11.84RR152 pKa = 11.84ILCAFWEE159 pKa = 4.4AMNVLYY165 pKa = 10.54KK166 pKa = 11.15DD167 pKa = 3.94NLDD170 pKa = 3.75SLALHH175 pKa = 6.48YY176 pKa = 10.92YY177 pKa = 10.54SLTSQCLSLRR187 pKa = 11.84TTAEE191 pKa = 3.79SSTVFALRR199 pKa = 11.84RR200 pKa = 11.84ALGMPDD206 pKa = 3.49PANSFHH212 pKa = 6.2QAVARR217 pKa = 11.84LHH219 pKa = 6.12CTTSVPYY226 pKa = 7.93TTTTFTHH233 pKa = 6.72KK234 pKa = 9.92PDD236 pKa = 4.36PPHH239 pKa = 6.59PRR241 pKa = 11.84PDD243 pKa = 4.7RR244 pKa = 11.84IHH246 pKa = 6.66CALYY250 pKa = 10.51VPPTNKK256 pKa = 10.17APVWEE261 pKa = 4.56TKK263 pKa = 7.35TEE265 pKa = 3.98RR266 pKa = 11.84LVWVSALVLLAAQGDD281 pKa = 4.23TALLAAQASFLAWTARR297 pKa = 11.84TIIPFF302 pKa = 3.75
MM1 pKa = 7.32SRR3 pKa = 11.84LMLTGVRR10 pKa = 11.84PSASPVSMHH19 pKa = 6.32TPAAHH24 pKa = 6.69CAPSSSAVVHH34 pKa = 5.82LHH36 pKa = 5.79SAAIHH41 pKa = 4.97MAKK44 pKa = 10.16AVSTCPGARR53 pKa = 11.84SKK55 pKa = 10.94AARR58 pKa = 11.84NSSLIGSTNRR68 pKa = 11.84LPTQPITSAHH78 pKa = 6.35IPRR81 pKa = 11.84TSGDD85 pKa = 3.14TSSPPNSSTILATYY99 pKa = 9.11KK100 pKa = 10.65ARR102 pKa = 11.84AISSNDD108 pKa = 2.51IPRR111 pKa = 11.84FYY113 pKa = 10.92GGRR116 pKa = 11.84YY117 pKa = 6.2TRR119 pKa = 11.84ARR121 pKa = 11.84EE122 pKa = 3.46QAQYY126 pKa = 10.65AKK128 pKa = 9.9HH129 pKa = 5.37GVGMPRR135 pKa = 11.84RR136 pKa = 11.84LRR138 pKa = 11.84AALWDD143 pKa = 3.66RR144 pKa = 11.84TVCSAHH150 pKa = 6.95RR151 pKa = 11.84RR152 pKa = 11.84ILCAFWEE159 pKa = 4.4AMNVLYY165 pKa = 10.54KK166 pKa = 11.15DD167 pKa = 3.94NLDD170 pKa = 3.75SLALHH175 pKa = 6.48YY176 pKa = 10.92YY177 pKa = 10.54SLTSQCLSLRR187 pKa = 11.84TTAEE191 pKa = 3.79SSTVFALRR199 pKa = 11.84RR200 pKa = 11.84ALGMPDD206 pKa = 3.49PANSFHH212 pKa = 6.2QAVARR217 pKa = 11.84LHH219 pKa = 6.12CTTSVPYY226 pKa = 7.93TTTTFTHH233 pKa = 6.72KK234 pKa = 9.92PDD236 pKa = 4.36PPHH239 pKa = 6.59PRR241 pKa = 11.84PDD243 pKa = 4.7RR244 pKa = 11.84IHH246 pKa = 6.66CALYY250 pKa = 10.51VPPTNKK256 pKa = 10.17APVWEE261 pKa = 4.56TKK263 pKa = 7.35TEE265 pKa = 3.98RR266 pKa = 11.84LVWVSALVLLAAQGDD281 pKa = 4.23TALLAAQASFLAWTARR297 pKa = 11.84TIIPFF302 pKa = 3.75
Molecular weight: 32.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
59898 |
110 |
3149 |
453.8 |
50.39 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.02 ± 0.18 | 3.257 ± 0.156 |
4.282 ± 0.133 | 5.164 ± 0.226 |
2.873 ± 0.114 | 5.748 ± 0.132 |
3.655 ± 0.096 | 3.706 ± 0.135 |
2.841 ± 0.114 | 9.585 ± 0.261 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.523 ± 0.1 | 3.251 ± 0.093 |
6.625 ± 0.442 | 3.78 ± 0.16 |
7.433 ± 0.163 | 6.611 ± 0.361 |
6.84 ± 0.179 | 6.92 ± 0.179 |
1.362 ± 0.074 | 3.524 ± 0.103 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
