
Blackberry leaf mottle associated virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Polyploviricotina; Ellioviricetes; Bunyavirales; Fimoviridae; Emaravirus; unclassified Emaravirus
Average proteome isoelectric point is 6.04
Get precalculated fractions of proteins
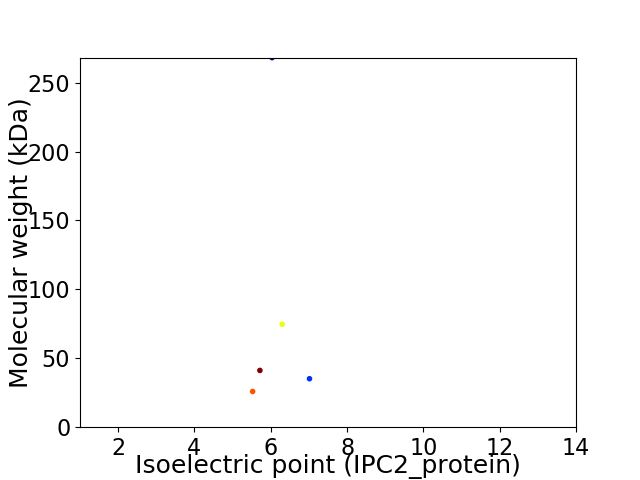
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1W5RWG7|A0A1W5RWG7_9VIRU Nucleocapsid protein OS=Blackberry leaf mottle associated virus OX=1966378 GN=P3 PE=4 SV=1
MM1 pKa = 7.06EE2 pKa = 6.01AKK4 pKa = 10.56AFDD7 pKa = 4.16YY8 pKa = 10.28TFHH11 pKa = 6.95SCTKK15 pKa = 9.55GGIMKK20 pKa = 10.39YY21 pKa = 9.99YY22 pKa = 10.85DD23 pKa = 4.4EE24 pKa = 6.3IDD26 pKa = 3.48FQLFFGFLEE35 pKa = 4.07EE36 pKa = 5.24LKK38 pKa = 10.99DD39 pKa = 4.29CIIYY43 pKa = 10.63DD44 pKa = 3.7HH45 pKa = 6.74LNKK48 pKa = 10.06LRR50 pKa = 11.84EE51 pKa = 4.1LAHH54 pKa = 6.75GDD56 pKa = 3.78LDD58 pKa = 4.33VSDD61 pKa = 4.42VDD63 pKa = 4.02GKK65 pKa = 9.53KK66 pKa = 8.15TAIQLTPDD74 pKa = 3.46FQTNQKK80 pKa = 10.14VMMYY84 pKa = 9.07FLRR87 pKa = 11.84YY88 pKa = 8.36HH89 pKa = 6.06QLLASLTGMKK99 pKa = 10.03PKK101 pKa = 10.69FIRR104 pKa = 11.84NIHH107 pKa = 6.31HH108 pKa = 7.34DD109 pKa = 3.23FHH111 pKa = 9.26IFGTLVRR118 pKa = 11.84GNEE121 pKa = 4.18MFPYY125 pKa = 10.14IPDD128 pKa = 3.49NMRR131 pKa = 11.84CLLLRR136 pKa = 11.84SDD138 pKa = 3.75ILKK141 pKa = 8.21TAVYY145 pKa = 9.8QVVKK149 pKa = 10.83GKK151 pKa = 10.76DD152 pKa = 3.09PMAALNMANGRR163 pKa = 11.84QPKK166 pKa = 9.19GFSYY170 pKa = 10.85KK171 pKa = 10.17EE172 pKa = 3.81QTSSYY177 pKa = 8.36TFQVIAISYY186 pKa = 9.06IALKK190 pKa = 10.59SSTEE194 pKa = 3.89IPGDD198 pKa = 3.43EE199 pKa = 4.31CFVPRR204 pKa = 11.84LVQKK208 pKa = 11.01SPDD211 pKa = 3.64VNDD214 pKa = 4.7DD215 pKa = 3.48EE216 pKa = 6.18SSGDD220 pKa = 3.58EE221 pKa = 4.13LNIIVV226 pKa = 4.35
MM1 pKa = 7.06EE2 pKa = 6.01AKK4 pKa = 10.56AFDD7 pKa = 4.16YY8 pKa = 10.28TFHH11 pKa = 6.95SCTKK15 pKa = 9.55GGIMKK20 pKa = 10.39YY21 pKa = 9.99YY22 pKa = 10.85DD23 pKa = 4.4EE24 pKa = 6.3IDD26 pKa = 3.48FQLFFGFLEE35 pKa = 4.07EE36 pKa = 5.24LKK38 pKa = 10.99DD39 pKa = 4.29CIIYY43 pKa = 10.63DD44 pKa = 3.7HH45 pKa = 6.74LNKK48 pKa = 10.06LRR50 pKa = 11.84EE51 pKa = 4.1LAHH54 pKa = 6.75GDD56 pKa = 3.78LDD58 pKa = 4.33VSDD61 pKa = 4.42VDD63 pKa = 4.02GKK65 pKa = 9.53KK66 pKa = 8.15TAIQLTPDD74 pKa = 3.46FQTNQKK80 pKa = 10.14VMMYY84 pKa = 9.07FLRR87 pKa = 11.84YY88 pKa = 8.36HH89 pKa = 6.06QLLASLTGMKK99 pKa = 10.03PKK101 pKa = 10.69FIRR104 pKa = 11.84NIHH107 pKa = 6.31HH108 pKa = 7.34DD109 pKa = 3.23FHH111 pKa = 9.26IFGTLVRR118 pKa = 11.84GNEE121 pKa = 4.18MFPYY125 pKa = 10.14IPDD128 pKa = 3.49NMRR131 pKa = 11.84CLLLRR136 pKa = 11.84SDD138 pKa = 3.75ILKK141 pKa = 8.21TAVYY145 pKa = 9.8QVVKK149 pKa = 10.83GKK151 pKa = 10.76DD152 pKa = 3.09PMAALNMANGRR163 pKa = 11.84QPKK166 pKa = 9.19GFSYY170 pKa = 10.85KK171 pKa = 10.17EE172 pKa = 3.81QTSSYY177 pKa = 8.36TFQVIAISYY186 pKa = 9.06IALKK190 pKa = 10.59SSTEE194 pKa = 3.89IPGDD198 pKa = 3.43EE199 pKa = 4.31CFVPRR204 pKa = 11.84LVQKK208 pKa = 11.01SPDD211 pKa = 3.64VNDD214 pKa = 4.7DD215 pKa = 3.48EE216 pKa = 6.18SSGDD220 pKa = 3.58EE221 pKa = 4.13LNIIVV226 pKa = 4.35
Molecular weight: 25.94 kDa
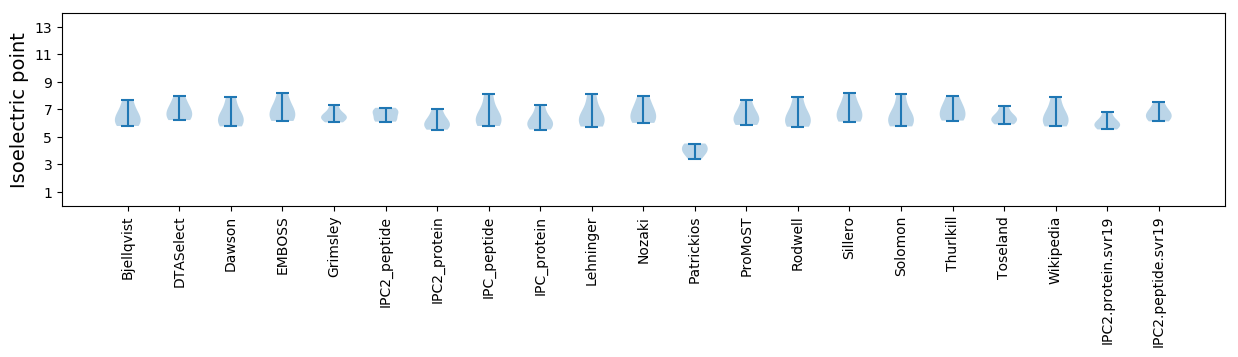
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1W5RYJ7|A0A1W5RYJ7_9VIRU Movement protein OS=Blackberry leaf mottle associated virus OX=1966378 GN=P4 PE=4 SV=1
MM1 pKa = 7.63PPKK4 pKa = 9.65TDD6 pKa = 3.28RR7 pKa = 11.84SASKK11 pKa = 9.16ATSSTPPSLVPANSLLVGSGTKK33 pKa = 10.26LRR35 pKa = 11.84TVKK38 pKa = 10.67LNNVVNKK45 pKa = 10.07VLLNPEE51 pKa = 3.93TSEE54 pKa = 3.99LKK56 pKa = 10.42PRR58 pKa = 11.84AKK60 pKa = 10.7DD61 pKa = 3.33PVPFTSASQHH71 pKa = 4.12QTNFTLTKK79 pKa = 10.36YY80 pKa = 10.39RR81 pKa = 11.84DD82 pKa = 3.76YY83 pKa = 11.85CNLTTVASYY92 pKa = 10.8LSRR95 pKa = 11.84SRR97 pKa = 11.84EE98 pKa = 4.11LKK100 pKa = 10.9DD101 pKa = 3.5EE102 pKa = 4.21LQKK105 pKa = 11.53SNLVLTTAAGKK116 pKa = 8.35TLTIVKK122 pKa = 9.67DD123 pKa = 3.93LNDD126 pKa = 3.72SDD128 pKa = 4.38VEE130 pKa = 4.32NVVSFNKK137 pKa = 10.18ACAIMSAGILKK148 pKa = 8.87HH149 pKa = 5.6TFLEE153 pKa = 4.51VFDD156 pKa = 4.05WTKK159 pKa = 10.97KK160 pKa = 10.27EE161 pKa = 4.01YY162 pKa = 10.92VQTTVEE168 pKa = 4.41GRR170 pKa = 11.84SAPDD174 pKa = 2.8ITIINKK180 pKa = 9.36LAGAMGLQPGNPYY193 pKa = 10.41YY194 pKa = 10.44WMIVPGYY201 pKa = 8.6EE202 pKa = 4.21FLYY205 pKa = 10.04EE206 pKa = 4.26LYY208 pKa = 9.64PAEE211 pKa = 4.09VLAYY215 pKa = 8.76TLVRR219 pKa = 11.84LEE221 pKa = 5.04FRR223 pKa = 11.84SQLNIPTDD231 pKa = 3.51MTDD234 pKa = 3.15ADD236 pKa = 4.48IISSLVMKK244 pKa = 9.26MNRR247 pKa = 11.84IHH249 pKa = 7.11ALEE252 pKa = 4.09STSFDD257 pKa = 3.28EE258 pKa = 5.57AINIVGEE265 pKa = 4.07DD266 pKa = 3.89NIRR269 pKa = 11.84EE270 pKa = 4.26VYY272 pKa = 10.68LEE274 pKa = 3.9LARR277 pKa = 11.84DD278 pKa = 3.45VGTTSKK284 pKa = 9.24TRR286 pKa = 11.84RR287 pKa = 11.84NDD289 pKa = 3.12EE290 pKa = 4.77AILKK294 pKa = 8.57FKK296 pKa = 10.81QLITNFGAALHH307 pKa = 5.71EE308 pKa = 4.17EE309 pKa = 4.46RR310 pKa = 11.84VRR312 pKa = 11.84AGHH315 pKa = 6.66AA316 pKa = 3.13
MM1 pKa = 7.63PPKK4 pKa = 9.65TDD6 pKa = 3.28RR7 pKa = 11.84SASKK11 pKa = 9.16ATSSTPPSLVPANSLLVGSGTKK33 pKa = 10.26LRR35 pKa = 11.84TVKK38 pKa = 10.67LNNVVNKK45 pKa = 10.07VLLNPEE51 pKa = 3.93TSEE54 pKa = 3.99LKK56 pKa = 10.42PRR58 pKa = 11.84AKK60 pKa = 10.7DD61 pKa = 3.33PVPFTSASQHH71 pKa = 4.12QTNFTLTKK79 pKa = 10.36YY80 pKa = 10.39RR81 pKa = 11.84DD82 pKa = 3.76YY83 pKa = 11.85CNLTTVASYY92 pKa = 10.8LSRR95 pKa = 11.84SRR97 pKa = 11.84EE98 pKa = 4.11LKK100 pKa = 10.9DD101 pKa = 3.5EE102 pKa = 4.21LQKK105 pKa = 11.53SNLVLTTAAGKK116 pKa = 8.35TLTIVKK122 pKa = 9.67DD123 pKa = 3.93LNDD126 pKa = 3.72SDD128 pKa = 4.38VEE130 pKa = 4.32NVVSFNKK137 pKa = 10.18ACAIMSAGILKK148 pKa = 8.87HH149 pKa = 5.6TFLEE153 pKa = 4.51VFDD156 pKa = 4.05WTKK159 pKa = 10.97KK160 pKa = 10.27EE161 pKa = 4.01YY162 pKa = 10.92VQTTVEE168 pKa = 4.41GRR170 pKa = 11.84SAPDD174 pKa = 2.8ITIINKK180 pKa = 9.36LAGAMGLQPGNPYY193 pKa = 10.41YY194 pKa = 10.44WMIVPGYY201 pKa = 8.6EE202 pKa = 4.21FLYY205 pKa = 10.04EE206 pKa = 4.26LYY208 pKa = 9.64PAEE211 pKa = 4.09VLAYY215 pKa = 8.76TLVRR219 pKa = 11.84LEE221 pKa = 5.04FRR223 pKa = 11.84SQLNIPTDD231 pKa = 3.51MTDD234 pKa = 3.15ADD236 pKa = 4.48IISSLVMKK244 pKa = 9.26MNRR247 pKa = 11.84IHH249 pKa = 7.11ALEE252 pKa = 4.09STSFDD257 pKa = 3.28EE258 pKa = 5.57AINIVGEE265 pKa = 4.07DD266 pKa = 3.89NIRR269 pKa = 11.84EE270 pKa = 4.26VYY272 pKa = 10.68LEE274 pKa = 3.9LARR277 pKa = 11.84DD278 pKa = 3.45VGTTSKK284 pKa = 9.24TRR286 pKa = 11.84RR287 pKa = 11.84NDD289 pKa = 3.12EE290 pKa = 4.77AILKK294 pKa = 8.57FKK296 pKa = 10.81QLITNFGAALHH307 pKa = 5.71EE308 pKa = 4.17EE309 pKa = 4.46RR310 pKa = 11.84VRR312 pKa = 11.84AGHH315 pKa = 6.66AA316 pKa = 3.13
Molecular weight: 35.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3859 |
226 |
2302 |
771.8 |
89.04 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.328 ± 0.624 | 1.71 ± 0.462 |
6.815 ± 0.624 | 5.649 ± 0.176 |
4.276 ± 0.231 | 3.421 ± 0.682 |
2.565 ± 0.187 | 8.526 ± 0.748 |
7.955 ± 0.197 | 9.692 ± 0.47 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.954 ± 0.438 | 6.867 ± 0.851 |
3.472 ± 0.221 | 2.799 ± 0.403 |
3.783 ± 0.388 | 7.1 ± 0.178 |
6.09 ± 0.513 | 5.96 ± 0.694 |
0.466 ± 0.128 | 5.571 ± 0.532 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
