
Breznakibacter xylanolyticus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Bacteroidia; Marinilabiliales; Marinilabiliaceae; Breznakibacter
Average proteome isoelectric point is 6.56
Get precalculated fractions of proteins
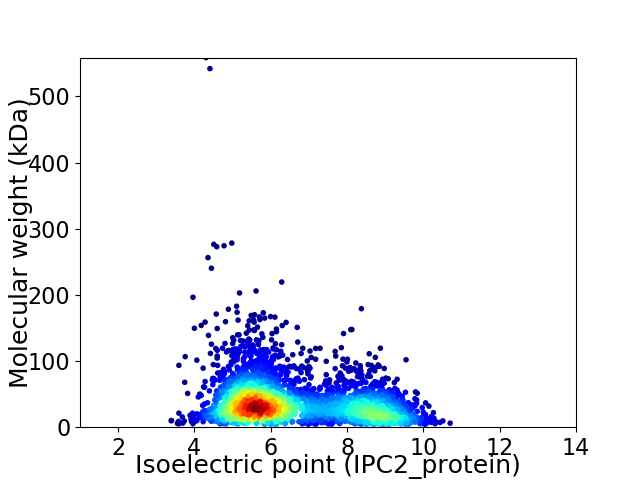
Virtual 2D-PAGE plot for 3475 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2W7MTE1|A0A2W7MTE1_9BACT Uncharacterized protein OS=Breznakibacter xylanolyticus OX=990 GN=LX69_03182 PE=4 SV=1
VV1 pKa = 7.59DD2 pKa = 4.14FQGALTVSVTVSDD15 pKa = 4.6GKK17 pKa = 9.99ATSAPYY23 pKa = 8.45QTLVTVTPVNDD34 pKa = 3.49VPVVEE39 pKa = 4.38EE40 pKa = 3.58DD41 pKa = 3.17HH42 pKa = 7.13YY43 pKa = 10.94YY44 pKa = 10.0TGEE47 pKa = 3.99NTSVNMAVLANDD59 pKa = 3.86HH60 pKa = 7.07DD61 pKa = 4.58LADD64 pKa = 5.3GINGGINPASLQIVQSAGHH83 pKa = 6.83GICLVLANGNIVYY96 pKa = 8.95TPYY99 pKa = 10.46PGFSGTDD106 pKa = 2.98QFTYY110 pKa = 10.27RR111 pKa = 11.84VSDD114 pKa = 3.06VGFPMPALSGTAKK127 pKa = 10.37VVVDD131 pKa = 4.2VARR134 pKa = 11.84MSPVCVADD142 pKa = 3.55AASVLEE148 pKa = 4.56DD149 pKa = 3.82EE150 pKa = 5.22FVDD153 pKa = 3.9IHH155 pKa = 7.03VLTNDD160 pKa = 3.48SDD162 pKa = 3.89SGGDD166 pKa = 3.15IDD168 pKa = 6.27ASTLSVFSHH177 pKa = 7.52PIHH180 pKa = 6.82GNASVQPNGVVRR192 pKa = 11.84YY193 pKa = 8.97RR194 pKa = 11.84PNVNYY199 pKa = 10.31HH200 pKa = 6.1GEE202 pKa = 4.09DD203 pKa = 3.17LFSYY207 pKa = 7.31TVRR210 pKa = 11.84DD211 pKa = 3.52LTGLIGNVGTVRR223 pKa = 11.84VTVLPVNDD231 pKa = 4.26APVALTGQFTTRR243 pKa = 11.84EE244 pKa = 4.16AVAVTIPLGQLASDD258 pKa = 4.23PDD260 pKa = 4.03GDD262 pKa = 4.24IDD264 pKa = 3.39WSRR267 pKa = 11.84FKK269 pKa = 10.48IVKK272 pKa = 9.61LPSSGTVSVDD282 pKa = 3.1VKK284 pKa = 11.17ALTVVYY290 pKa = 9.86RR291 pKa = 11.84PNTAYY296 pKa = 10.61SGADD300 pKa = 3.01LFEE303 pKa = 4.15FTVVDD308 pKa = 3.37SHH310 pKa = 6.65GVQSNRR316 pKa = 11.84GSAAISVSNEE326 pKa = 3.38SPMVVDD332 pKa = 5.16DD333 pKa = 4.14VGEE336 pKa = 4.11VAEE339 pKa = 5.07DD340 pKa = 3.31SSIALDD346 pKa = 3.75VMSNDD351 pKa = 4.0SDD353 pKa = 4.07LQGDD357 pKa = 4.3LDD359 pKa = 4.25GASLMIIVPPAHH371 pKa = 7.05GIASVDD377 pKa = 3.51ARR379 pKa = 11.84SHH381 pKa = 5.64LVNYY385 pKa = 9.62YY386 pKa = 9.6PALNYY391 pKa = 10.72YY392 pKa = 10.29GVDD395 pKa = 3.08EE396 pKa = 5.47FSYY399 pKa = 9.81QVCDD403 pKa = 3.26NDD405 pKa = 5.06GYY407 pKa = 10.89CGRR410 pKa = 11.84ATVRR414 pKa = 11.84LTVHH418 pKa = 6.82PVNDD422 pKa = 3.99APLALDD428 pKa = 5.09DD429 pKa = 5.36FFEE432 pKa = 5.95LDD434 pKa = 3.5EE435 pKa = 5.29DD436 pKa = 3.99EE437 pKa = 5.01SLLVDD442 pKa = 3.99VLLNDD447 pKa = 4.01SDD449 pKa = 5.55GDD451 pKa = 4.01GQLDD455 pKa = 3.48AGSISVVRR463 pKa = 11.84QPAHH467 pKa = 5.42GQVVVDD473 pKa = 4.26AQHH476 pKa = 6.16GRR478 pKa = 11.84LLVVLDD484 pKa = 3.91NDD486 pKa = 4.06FFGDD490 pKa = 3.77DD491 pKa = 3.28EE492 pKa = 4.38LTYY495 pKa = 10.28RR496 pKa = 11.84VCDD499 pKa = 3.79DD500 pKa = 5.09RR501 pKa = 11.84GACSQAVAYY510 pKa = 10.24LKK512 pKa = 10.43VRR514 pKa = 11.84PVNDD518 pKa = 3.15APVAVNDD525 pKa = 4.04KK526 pKa = 10.95LVVEE530 pKa = 4.06TDD532 pKa = 3.03EE533 pKa = 4.99RR534 pKa = 11.84VVFTPLDD541 pKa = 3.74NDD543 pKa = 3.75SDD545 pKa = 4.15VDD547 pKa = 4.17DD548 pKa = 4.92GLNLSTLTLIVAPTWGKK565 pKa = 11.0AEE567 pKa = 3.99VLGNGQLSYY576 pKa = 11.12QSDD579 pKa = 3.38AGYY582 pKa = 10.81VGVVTLRR589 pKa = 11.84YY590 pKa = 9.12RR591 pKa = 11.84VCDD594 pKa = 3.52VGGLCAQADD603 pKa = 3.83VEE605 pKa = 4.58INVVSGNQPPVAVGDD620 pKa = 3.81LFSVNEE626 pKa = 4.45DD627 pKa = 2.62QDD629 pKa = 3.19MWLSVLDD636 pKa = 3.83NDD638 pKa = 4.9FDD640 pKa = 4.93SRR642 pKa = 11.84DD643 pKa = 3.71HH644 pKa = 6.9LLLSSLTVVEE654 pKa = 4.86APLHH658 pKa = 5.26GWATVDD664 pKa = 3.42ASAGKK669 pKa = 10.01IIYY672 pKa = 10.22RR673 pKa = 11.84PDD675 pKa = 3.23ADD677 pKa = 3.72YY678 pKa = 10.99WGVDD682 pKa = 3.11YY683 pKa = 11.15LRR685 pKa = 11.84YY686 pKa = 9.58KK687 pKa = 10.57VCDD690 pKa = 4.14DD691 pKa = 5.19NILPLCAEE699 pKa = 4.29ASVSISVMPVNDD711 pKa = 4.11APVGHH716 pKa = 7.4ADD718 pKa = 3.02HH719 pKa = 7.0YY720 pKa = 10.63EE721 pKa = 3.98AFDD724 pKa = 3.38VGEE727 pKa = 4.28YY728 pKa = 9.65NWNVLINDD736 pKa = 3.88TDD738 pKa = 3.64VDD740 pKa = 4.12DD741 pKa = 4.66TKK743 pKa = 11.21LSCRR747 pKa = 11.84LVDD750 pKa = 3.68ASGIAGEE757 pKa = 4.53VTLSAQGWLRR767 pKa = 11.84WRR769 pKa = 11.84PLPGCYY775 pKa = 9.74CSVQSLMYY783 pKa = 8.94EE784 pKa = 4.1VCDD787 pKa = 3.63PAGEE791 pKa = 4.18CDD793 pKa = 3.74VVQVLIDD800 pKa = 3.6VQYY803 pKa = 11.35SDD805 pKa = 3.86WDD807 pKa = 3.6GDD809 pKa = 4.34GIPDD813 pKa = 4.51AVDD816 pKa = 3.35GDD818 pKa = 4.16GDD820 pKa = 4.12SDD822 pKa = 4.97GDD824 pKa = 3.81GLPNYY829 pKa = 9.83RR830 pKa = 11.84DD831 pKa = 3.57ADD833 pKa = 3.85SDD835 pKa = 3.93GDD837 pKa = 4.47GIDD840 pKa = 3.61DD841 pKa = 4.34TIEE844 pKa = 4.21GGVADD849 pKa = 4.45VCTQFPVDD857 pKa = 3.76SDD859 pKa = 3.89GDD861 pKa = 4.13GVPDD865 pKa = 5.33FLDD868 pKa = 4.08ADD870 pKa = 4.07ADD872 pKa = 4.14GDD874 pKa = 4.3GVPDD878 pKa = 3.97AVEE881 pKa = 4.24LTDD884 pKa = 5.29DD885 pKa = 4.43CDD887 pKa = 4.64GDD889 pKa = 4.61GIPNHH894 pKa = 6.87LDD896 pKa = 3.47VVDD899 pKa = 4.54DD900 pKa = 3.69CTNRR904 pKa = 11.84VLVPDD909 pKa = 4.35TFTPNGDD916 pKa = 3.39GVNDD920 pKa = 3.66RR921 pKa = 11.84FVIPIVRR928 pKa = 11.84EE929 pKa = 3.75YY930 pKa = 10.24PINRR934 pKa = 11.84LTIFNRR940 pKa = 11.84WGNQVYY946 pKa = 9.58VADD949 pKa = 4.5NYY951 pKa = 11.14RR952 pKa = 11.84NDD954 pKa = 3.21WDD956 pKa = 4.13GRR958 pKa = 11.84SSTVTMGTNVLPEE971 pKa = 3.73GTYY974 pKa = 10.2FYY976 pKa = 11.4VLTLGVEE983 pKa = 4.14SRR985 pKa = 11.84LMKK988 pKa = 10.73GIVYY992 pKa = 10.09LKK994 pKa = 10.55KK995 pKa = 10.68
VV1 pKa = 7.59DD2 pKa = 4.14FQGALTVSVTVSDD15 pKa = 4.6GKK17 pKa = 9.99ATSAPYY23 pKa = 8.45QTLVTVTPVNDD34 pKa = 3.49VPVVEE39 pKa = 4.38EE40 pKa = 3.58DD41 pKa = 3.17HH42 pKa = 7.13YY43 pKa = 10.94YY44 pKa = 10.0TGEE47 pKa = 3.99NTSVNMAVLANDD59 pKa = 3.86HH60 pKa = 7.07DD61 pKa = 4.58LADD64 pKa = 5.3GINGGINPASLQIVQSAGHH83 pKa = 6.83GICLVLANGNIVYY96 pKa = 8.95TPYY99 pKa = 10.46PGFSGTDD106 pKa = 2.98QFTYY110 pKa = 10.27RR111 pKa = 11.84VSDD114 pKa = 3.06VGFPMPALSGTAKK127 pKa = 10.37VVVDD131 pKa = 4.2VARR134 pKa = 11.84MSPVCVADD142 pKa = 3.55AASVLEE148 pKa = 4.56DD149 pKa = 3.82EE150 pKa = 5.22FVDD153 pKa = 3.9IHH155 pKa = 7.03VLTNDD160 pKa = 3.48SDD162 pKa = 3.89SGGDD166 pKa = 3.15IDD168 pKa = 6.27ASTLSVFSHH177 pKa = 7.52PIHH180 pKa = 6.82GNASVQPNGVVRR192 pKa = 11.84YY193 pKa = 8.97RR194 pKa = 11.84PNVNYY199 pKa = 10.31HH200 pKa = 6.1GEE202 pKa = 4.09DD203 pKa = 3.17LFSYY207 pKa = 7.31TVRR210 pKa = 11.84DD211 pKa = 3.52LTGLIGNVGTVRR223 pKa = 11.84VTVLPVNDD231 pKa = 4.26APVALTGQFTTRR243 pKa = 11.84EE244 pKa = 4.16AVAVTIPLGQLASDD258 pKa = 4.23PDD260 pKa = 4.03GDD262 pKa = 4.24IDD264 pKa = 3.39WSRR267 pKa = 11.84FKK269 pKa = 10.48IVKK272 pKa = 9.61LPSSGTVSVDD282 pKa = 3.1VKK284 pKa = 11.17ALTVVYY290 pKa = 9.86RR291 pKa = 11.84PNTAYY296 pKa = 10.61SGADD300 pKa = 3.01LFEE303 pKa = 4.15FTVVDD308 pKa = 3.37SHH310 pKa = 6.65GVQSNRR316 pKa = 11.84GSAAISVSNEE326 pKa = 3.38SPMVVDD332 pKa = 5.16DD333 pKa = 4.14VGEE336 pKa = 4.11VAEE339 pKa = 5.07DD340 pKa = 3.31SSIALDD346 pKa = 3.75VMSNDD351 pKa = 4.0SDD353 pKa = 4.07LQGDD357 pKa = 4.3LDD359 pKa = 4.25GASLMIIVPPAHH371 pKa = 7.05GIASVDD377 pKa = 3.51ARR379 pKa = 11.84SHH381 pKa = 5.64LVNYY385 pKa = 9.62YY386 pKa = 9.6PALNYY391 pKa = 10.72YY392 pKa = 10.29GVDD395 pKa = 3.08EE396 pKa = 5.47FSYY399 pKa = 9.81QVCDD403 pKa = 3.26NDD405 pKa = 5.06GYY407 pKa = 10.89CGRR410 pKa = 11.84ATVRR414 pKa = 11.84LTVHH418 pKa = 6.82PVNDD422 pKa = 3.99APLALDD428 pKa = 5.09DD429 pKa = 5.36FFEE432 pKa = 5.95LDD434 pKa = 3.5EE435 pKa = 5.29DD436 pKa = 3.99EE437 pKa = 5.01SLLVDD442 pKa = 3.99VLLNDD447 pKa = 4.01SDD449 pKa = 5.55GDD451 pKa = 4.01GQLDD455 pKa = 3.48AGSISVVRR463 pKa = 11.84QPAHH467 pKa = 5.42GQVVVDD473 pKa = 4.26AQHH476 pKa = 6.16GRR478 pKa = 11.84LLVVLDD484 pKa = 3.91NDD486 pKa = 4.06FFGDD490 pKa = 3.77DD491 pKa = 3.28EE492 pKa = 4.38LTYY495 pKa = 10.28RR496 pKa = 11.84VCDD499 pKa = 3.79DD500 pKa = 5.09RR501 pKa = 11.84GACSQAVAYY510 pKa = 10.24LKK512 pKa = 10.43VRR514 pKa = 11.84PVNDD518 pKa = 3.15APVAVNDD525 pKa = 4.04KK526 pKa = 10.95LVVEE530 pKa = 4.06TDD532 pKa = 3.03EE533 pKa = 4.99RR534 pKa = 11.84VVFTPLDD541 pKa = 3.74NDD543 pKa = 3.75SDD545 pKa = 4.15VDD547 pKa = 4.17DD548 pKa = 4.92GLNLSTLTLIVAPTWGKK565 pKa = 11.0AEE567 pKa = 3.99VLGNGQLSYY576 pKa = 11.12QSDD579 pKa = 3.38AGYY582 pKa = 10.81VGVVTLRR589 pKa = 11.84YY590 pKa = 9.12RR591 pKa = 11.84VCDD594 pKa = 3.52VGGLCAQADD603 pKa = 3.83VEE605 pKa = 4.58INVVSGNQPPVAVGDD620 pKa = 3.81LFSVNEE626 pKa = 4.45DD627 pKa = 2.62QDD629 pKa = 3.19MWLSVLDD636 pKa = 3.83NDD638 pKa = 4.9FDD640 pKa = 4.93SRR642 pKa = 11.84DD643 pKa = 3.71HH644 pKa = 6.9LLLSSLTVVEE654 pKa = 4.86APLHH658 pKa = 5.26GWATVDD664 pKa = 3.42ASAGKK669 pKa = 10.01IIYY672 pKa = 10.22RR673 pKa = 11.84PDD675 pKa = 3.23ADD677 pKa = 3.72YY678 pKa = 10.99WGVDD682 pKa = 3.11YY683 pKa = 11.15LRR685 pKa = 11.84YY686 pKa = 9.58KK687 pKa = 10.57VCDD690 pKa = 4.14DD691 pKa = 5.19NILPLCAEE699 pKa = 4.29ASVSISVMPVNDD711 pKa = 4.11APVGHH716 pKa = 7.4ADD718 pKa = 3.02HH719 pKa = 7.0YY720 pKa = 10.63EE721 pKa = 3.98AFDD724 pKa = 3.38VGEE727 pKa = 4.28YY728 pKa = 9.65NWNVLINDD736 pKa = 3.88TDD738 pKa = 3.64VDD740 pKa = 4.12DD741 pKa = 4.66TKK743 pKa = 11.21LSCRR747 pKa = 11.84LVDD750 pKa = 3.68ASGIAGEE757 pKa = 4.53VTLSAQGWLRR767 pKa = 11.84WRR769 pKa = 11.84PLPGCYY775 pKa = 9.74CSVQSLMYY783 pKa = 8.94EE784 pKa = 4.1VCDD787 pKa = 3.63PAGEE791 pKa = 4.18CDD793 pKa = 3.74VVQVLIDD800 pKa = 3.6VQYY803 pKa = 11.35SDD805 pKa = 3.86WDD807 pKa = 3.6GDD809 pKa = 4.34GIPDD813 pKa = 4.51AVDD816 pKa = 3.35GDD818 pKa = 4.16GDD820 pKa = 4.12SDD822 pKa = 4.97GDD824 pKa = 3.81GLPNYY829 pKa = 9.83RR830 pKa = 11.84DD831 pKa = 3.57ADD833 pKa = 3.85SDD835 pKa = 3.93GDD837 pKa = 4.47GIDD840 pKa = 3.61DD841 pKa = 4.34TIEE844 pKa = 4.21GGVADD849 pKa = 4.45VCTQFPVDD857 pKa = 3.76SDD859 pKa = 3.89GDD861 pKa = 4.13GVPDD865 pKa = 5.33FLDD868 pKa = 4.08ADD870 pKa = 4.07ADD872 pKa = 4.14GDD874 pKa = 4.3GVPDD878 pKa = 3.97AVEE881 pKa = 4.24LTDD884 pKa = 5.29DD885 pKa = 4.43CDD887 pKa = 4.64GDD889 pKa = 4.61GIPNHH894 pKa = 6.87LDD896 pKa = 3.47VVDD899 pKa = 4.54DD900 pKa = 3.69CTNRR904 pKa = 11.84VLVPDD909 pKa = 4.35TFTPNGDD916 pKa = 3.39GVNDD920 pKa = 3.66RR921 pKa = 11.84FVIPIVRR928 pKa = 11.84EE929 pKa = 3.75YY930 pKa = 10.24PINRR934 pKa = 11.84LTIFNRR940 pKa = 11.84WGNQVYY946 pKa = 9.58VADD949 pKa = 4.5NYY951 pKa = 11.14RR952 pKa = 11.84NDD954 pKa = 3.21WDD956 pKa = 4.13GRR958 pKa = 11.84SSTVTMGTNVLPEE971 pKa = 3.73GTYY974 pKa = 10.2FYY976 pKa = 11.4VLTLGVEE983 pKa = 4.14SRR985 pKa = 11.84LMKK988 pKa = 10.73GIVYY992 pKa = 10.09LKK994 pKa = 10.55KK995 pKa = 10.68
Molecular weight: 106.62 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2W7NS53|A0A2W7NS53_9BACT Uncharacterized protein OS=Breznakibacter xylanolyticus OX=990 GN=LX69_01980 PE=4 SV=1
MM1 pKa = 7.47FYY3 pKa = 11.42SMFLICKK10 pKa = 9.64RR11 pKa = 11.84NRR13 pKa = 11.84NRR15 pKa = 11.84HH16 pKa = 4.94RR17 pKa = 11.84KK18 pKa = 8.82RR19 pKa = 11.84LRR21 pKa = 11.84KK22 pKa = 9.5DD23 pKa = 2.81RR24 pKa = 11.84CKK26 pKa = 10.93LLIPTVKK33 pKa = 10.42NCLLSQFYY41 pKa = 10.5AIALSCPSEE50 pKa = 3.97
MM1 pKa = 7.47FYY3 pKa = 11.42SMFLICKK10 pKa = 9.64RR11 pKa = 11.84NRR13 pKa = 11.84NRR15 pKa = 11.84HH16 pKa = 4.94RR17 pKa = 11.84KK18 pKa = 8.82RR19 pKa = 11.84LRR21 pKa = 11.84KK22 pKa = 9.5DD23 pKa = 2.81RR24 pKa = 11.84CKK26 pKa = 10.93LLIPTVKK33 pKa = 10.42NCLLSQFYY41 pKa = 10.5AIALSCPSEE50 pKa = 3.97
Molecular weight: 6.06 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1239837 |
31 |
5336 |
356.8 |
40.02 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.368 ± 0.044 | 1.026 ± 0.017 |
5.54 ± 0.026 | 5.752 ± 0.045 |
4.618 ± 0.03 | 6.91 ± 0.044 |
2.323 ± 0.023 | 6.709 ± 0.042 |
5.791 ± 0.05 | 9.353 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.817 ± 0.025 | 5.176 ± 0.049 |
3.891 ± 0.03 | 3.903 ± 0.025 |
4.659 ± 0.038 | 6.358 ± 0.049 |
5.736 ± 0.055 | 6.998 ± 0.041 |
1.265 ± 0.016 | 3.805 ± 0.026 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
