
Cebus albifrons polyomavirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Sepolyvirales; Polyomaviridae; Betapolyomavirus
Average proteome isoelectric point is 5.79
Get precalculated fractions of proteins
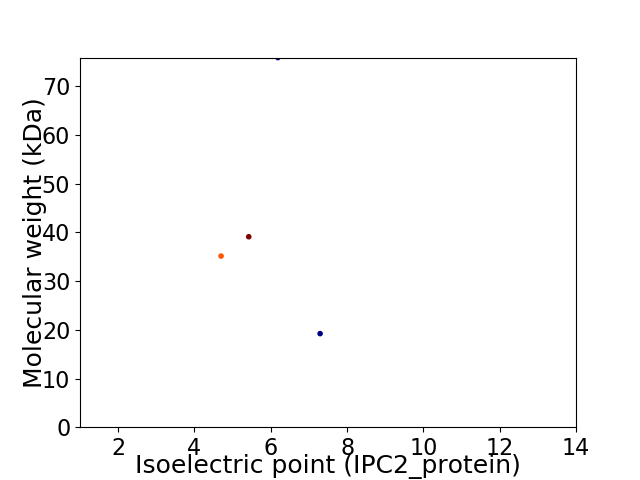
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|K7QLH5|K7QLH5_9POLY Minor capsid protein OS=Cebus albifrons polyomavirus 1 OX=1236392 PE=3 SV=1
MM1 pKa = 7.12GAVLAVLAEE10 pKa = 4.16VFEE13 pKa = 4.57LASVTGLSVDD23 pKa = 4.64AILSGEE29 pKa = 4.2AFTTAEE35 pKa = 4.35LLQAHH40 pKa = 7.43IANLVTVGGLTEE52 pKa = 4.49AEE54 pKa = 3.83ALAATEE60 pKa = 4.37VTAEE64 pKa = 4.26AYY66 pKa = 9.13AALTSITSTFPQAFAALAATEE87 pKa = 4.1LTATGSLTVGAAVAAALYY105 pKa = 8.99PYY107 pKa = 9.42TYY109 pKa = 11.04DD110 pKa = 4.21HH111 pKa = 6.41STPLANLNTNMALQVWIPDD130 pKa = 3.65LDD132 pKa = 4.16LDD134 pKa = 4.32FPGVRR139 pKa = 11.84PFVRR143 pKa = 11.84FVNYY147 pKa = 9.59IDD149 pKa = 3.91PAQWATDD156 pKa = 3.61LFQAVGRR163 pKa = 11.84YY164 pKa = 7.31FWQTAQRR171 pKa = 11.84YY172 pKa = 5.77GQNLIEE178 pKa = 4.6HH179 pKa = 6.88EE180 pKa = 4.09LRR182 pKa = 11.84EE183 pKa = 4.28TSRR186 pKa = 11.84EE187 pKa = 4.0LAVTAVTSVSDD198 pKa = 3.1VLARR202 pKa = 11.84YY203 pKa = 9.57FEE205 pKa = 4.07TARR208 pKa = 11.84WAVSLLPRR216 pKa = 11.84NIYY219 pKa = 10.11QSLQNYY225 pKa = 6.9YY226 pKa = 10.53QEE228 pKa = 5.27LPPLNPIQVRR238 pKa = 11.84QLQRR242 pKa = 11.84RR243 pKa = 11.84IGGTIPDD250 pKa = 4.12RR251 pKa = 11.84LTFHH255 pKa = 7.14DD256 pKa = 4.92TIQTAEE262 pKa = 4.16YY263 pKa = 9.86VEE265 pKa = 4.43KK266 pKa = 11.24AEE268 pKa = 4.63APGGANQRR276 pKa = 11.84VAHH279 pKa = 6.83DD280 pKa = 3.62WLLPLLLGLYY290 pKa = 10.75GDD292 pKa = 5.13ISPSWEE298 pKa = 3.97STLEE302 pKa = 4.04DD303 pKa = 4.22LEE305 pKa = 4.52EE306 pKa = 5.02EE307 pKa = 4.01EE308 pKa = 6.13DD309 pKa = 3.83APQKK313 pKa = 10.67KK314 pKa = 9.32KK315 pKa = 10.48RR316 pKa = 11.84KK317 pKa = 8.98KK318 pKa = 10.4SKK320 pKa = 10.52
MM1 pKa = 7.12GAVLAVLAEE10 pKa = 4.16VFEE13 pKa = 4.57LASVTGLSVDD23 pKa = 4.64AILSGEE29 pKa = 4.2AFTTAEE35 pKa = 4.35LLQAHH40 pKa = 7.43IANLVTVGGLTEE52 pKa = 4.49AEE54 pKa = 3.83ALAATEE60 pKa = 4.37VTAEE64 pKa = 4.26AYY66 pKa = 9.13AALTSITSTFPQAFAALAATEE87 pKa = 4.1LTATGSLTVGAAVAAALYY105 pKa = 8.99PYY107 pKa = 9.42TYY109 pKa = 11.04DD110 pKa = 4.21HH111 pKa = 6.41STPLANLNTNMALQVWIPDD130 pKa = 3.65LDD132 pKa = 4.16LDD134 pKa = 4.32FPGVRR139 pKa = 11.84PFVRR143 pKa = 11.84FVNYY147 pKa = 9.59IDD149 pKa = 3.91PAQWATDD156 pKa = 3.61LFQAVGRR163 pKa = 11.84YY164 pKa = 7.31FWQTAQRR171 pKa = 11.84YY172 pKa = 5.77GQNLIEE178 pKa = 4.6HH179 pKa = 6.88EE180 pKa = 4.09LRR182 pKa = 11.84EE183 pKa = 4.28TSRR186 pKa = 11.84EE187 pKa = 4.0LAVTAVTSVSDD198 pKa = 3.1VLARR202 pKa = 11.84YY203 pKa = 9.57FEE205 pKa = 4.07TARR208 pKa = 11.84WAVSLLPRR216 pKa = 11.84NIYY219 pKa = 10.11QSLQNYY225 pKa = 6.9YY226 pKa = 10.53QEE228 pKa = 5.27LPPLNPIQVRR238 pKa = 11.84QLQRR242 pKa = 11.84RR243 pKa = 11.84IGGTIPDD250 pKa = 4.12RR251 pKa = 11.84LTFHH255 pKa = 7.14DD256 pKa = 4.92TIQTAEE262 pKa = 4.16YY263 pKa = 9.86VEE265 pKa = 4.43KK266 pKa = 11.24AEE268 pKa = 4.63APGGANQRR276 pKa = 11.84VAHH279 pKa = 6.83DD280 pKa = 3.62WLLPLLLGLYY290 pKa = 10.75GDD292 pKa = 5.13ISPSWEE298 pKa = 3.97STLEE302 pKa = 4.04DD303 pKa = 4.22LEE305 pKa = 4.52EE306 pKa = 5.02EE307 pKa = 4.01EE308 pKa = 6.13DD309 pKa = 3.83APQKK313 pKa = 10.67KK314 pKa = 9.32KK315 pKa = 10.48RR316 pKa = 11.84KK317 pKa = 8.98KK318 pKa = 10.4SKK320 pKa = 10.52
Molecular weight: 35.15 kDa
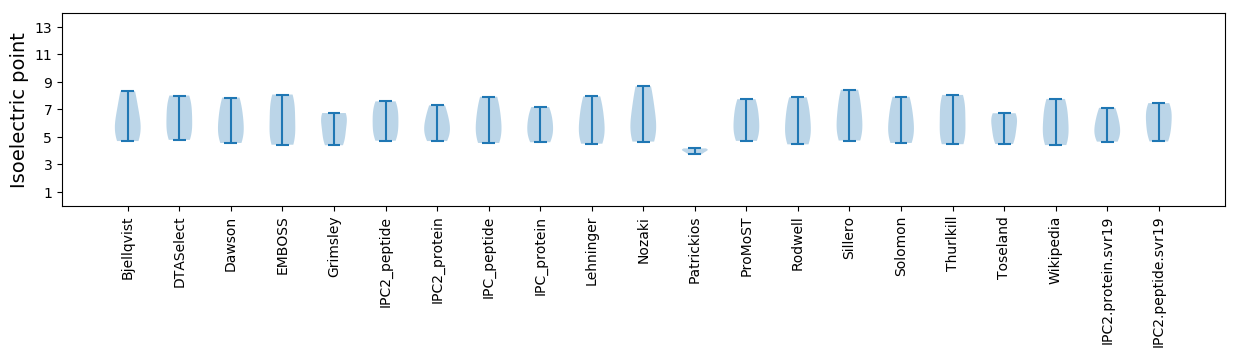
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|K7QLH5|K7QLH5_9POLY Minor capsid protein OS=Cebus albifrons polyomavirus 1 OX=1236392 PE=3 SV=1
MM1 pKa = 7.89DD2 pKa = 3.6HH3 pKa = 6.91TLTRR7 pKa = 11.84DD8 pKa = 3.26EE9 pKa = 5.01SKK11 pKa = 11.4LLMEE15 pKa = 5.29LLNLPMEE22 pKa = 4.45QYY24 pKa = 11.86GNFPLMRR31 pKa = 11.84KK32 pKa = 9.49AFLQKK37 pKa = 10.55CKK39 pKa = 10.03ILHH42 pKa = 6.38PDD44 pKa = 2.98KK45 pKa = 11.56GGDD48 pKa = 3.43QEE50 pKa = 4.41TAKK53 pKa = 10.37MLISLYY59 pKa = 10.35KK60 pKa = 9.96RR61 pKa = 11.84LEE63 pKa = 4.18AEE65 pKa = 4.28VQSLNTDD72 pKa = 3.4DD73 pKa = 6.37CFTTDD78 pKa = 3.41QVCEE82 pKa = 3.83ISNMIFLKK90 pKa = 10.43DD91 pKa = 3.06WRR93 pKa = 11.84TCRR96 pKa = 11.84MGSVKK101 pKa = 9.31CTCLFCLLKK110 pKa = 10.49RR111 pKa = 11.84EE112 pKa = 4.74HH113 pKa = 6.19KK114 pKa = 9.7QQLIGRR120 pKa = 11.84PKK122 pKa = 9.67VWGTCYY128 pKa = 10.5CFNCYY133 pKa = 9.79ILWFGLEE140 pKa = 3.73HH141 pKa = 6.69TYY143 pKa = 10.86DD144 pKa = 4.34IFLSWKK150 pKa = 9.6ALIAITPFRR159 pKa = 11.84NLNII163 pKa = 4.04
MM1 pKa = 7.89DD2 pKa = 3.6HH3 pKa = 6.91TLTRR7 pKa = 11.84DD8 pKa = 3.26EE9 pKa = 5.01SKK11 pKa = 11.4LLMEE15 pKa = 5.29LLNLPMEE22 pKa = 4.45QYY24 pKa = 11.86GNFPLMRR31 pKa = 11.84KK32 pKa = 9.49AFLQKK37 pKa = 10.55CKK39 pKa = 10.03ILHH42 pKa = 6.38PDD44 pKa = 2.98KK45 pKa = 11.56GGDD48 pKa = 3.43QEE50 pKa = 4.41TAKK53 pKa = 10.37MLISLYY59 pKa = 10.35KK60 pKa = 9.96RR61 pKa = 11.84LEE63 pKa = 4.18AEE65 pKa = 4.28VQSLNTDD72 pKa = 3.4DD73 pKa = 6.37CFTTDD78 pKa = 3.41QVCEE82 pKa = 3.83ISNMIFLKK90 pKa = 10.43DD91 pKa = 3.06WRR93 pKa = 11.84TCRR96 pKa = 11.84MGSVKK101 pKa = 9.31CTCLFCLLKK110 pKa = 10.49RR111 pKa = 11.84EE112 pKa = 4.74HH113 pKa = 6.19KK114 pKa = 9.7QQLIGRR120 pKa = 11.84PKK122 pKa = 9.67VWGTCYY128 pKa = 10.5CFNCYY133 pKa = 9.79ILWFGLEE140 pKa = 3.73HH141 pKa = 6.69TYY143 pKa = 10.86DD144 pKa = 4.34IFLSWKK150 pKa = 9.6ALIAITPFRR159 pKa = 11.84NLNII163 pKa = 4.04
Molecular weight: 19.22 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1495 |
163 |
659 |
373.8 |
42.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.421 ± 2.057 | 2.408 ± 0.919 |
5.351 ± 0.424 | 6.756 ± 0.259 |
5.151 ± 0.594 | 5.217 ± 0.808 |
1.873 ± 0.366 | 4.95 ± 0.342 |
7.09 ± 1.69 | 11.037 ± 0.891 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.14 ± 0.483 | 4.415 ± 0.406 |
5.217 ± 1.132 | 4.615 ± 0.281 |
3.612 ± 0.569 | 5.619 ± 0.399 |
7.157 ± 0.528 | 5.953 ± 0.851 |
1.405 ± 0.239 | 3.612 ± 0.139 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
