
Pseudomonas knackmussii (strain DSM 6978 / LMG 23759 / B13)
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Pseudomonadales; Pseudomonadaceae; Pseudomonas; Pseudomonas knackmussii
Average proteome isoelectric point is 6.41
Get precalculated fractions of proteins
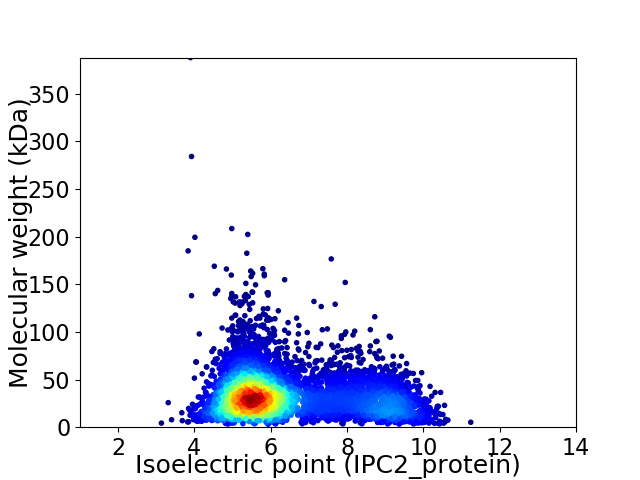
Virtual 2D-PAGE plot for 5616 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A024HG58|A0A024HG58_PSEKB Uncharacterized protein OS=Pseudomonas knackmussii (strain DSM 6978 / LMG 23759 / B13) OX=1301098 GN=PKB_2140 PE=4 SV=1
MM1 pKa = 7.37FRR3 pKa = 11.84LTPISAALLLLAAGQAMADD22 pKa = 3.78DD23 pKa = 4.36SSANQVQNGEE33 pKa = 4.05QNIANVSQKK42 pKa = 9.46QASWSTANQTQTGTGNDD59 pKa = 3.13AMAVQQEE66 pKa = 4.58STGSIVQSLSGDD78 pKa = 3.15HH79 pKa = 5.93NAAYY83 pKa = 10.34GEE85 pKa = 4.11QNYY88 pKa = 10.38GEE90 pKa = 4.5INSSINQQGNGDD102 pKa = 3.72WNDD105 pKa = 3.4NFASQYY111 pKa = 10.45YY112 pKa = 10.56GSDD115 pKa = 3.26NQALQIQDD123 pKa = 3.37GSRR126 pKa = 11.84SFSMVYY132 pKa = 9.68QEE134 pKa = 4.66GQVGSQGTTQQSGGNHH150 pKa = 5.71EE151 pKa = 4.95ANIEE155 pKa = 4.01QIGFGEE161 pKa = 4.55GNRR164 pKa = 11.84ALIIQSGDD172 pKa = 3.17ANRR175 pKa = 11.84GSIKK179 pKa = 10.53QSDD182 pKa = 4.35QISGHH187 pKa = 4.93QEE189 pKa = 3.71VYY191 pKa = 10.25QQGSSNTANGWQNLGLGGTVLFNQAGDD218 pKa = 3.54NNYY221 pKa = 10.7AEE223 pKa = 4.67VWQDD227 pKa = 3.45SQDD230 pKa = 3.33SSQASIAQFGSTNEE244 pKa = 3.86AVIDD248 pKa = 3.84QSFGMAMDD256 pKa = 4.91PYY258 pKa = 11.13GVGNVASINQSGDD271 pKa = 3.17RR272 pKa = 11.84NAAWADD278 pKa = 3.47QYY280 pKa = 11.4EE281 pKa = 4.83SKK283 pKa = 11.02GSNTALYY290 pKa = 10.74QNGTDD295 pKa = 4.02NLQLTYY301 pKa = 9.66QTGSGHH307 pKa = 5.71SLVVTSNGTGNKK319 pKa = 9.52VYY321 pKa = 10.47MSDD324 pKa = 2.79WQGARR329 pKa = 11.84RR330 pKa = 11.84GGQSGQGNNAAIAQNGGGNVAQVKK354 pKa = 9.1QQDD357 pKa = 4.04TGNKK361 pKa = 7.52TQLTQNGTNNDD372 pKa = 3.8LNVSQAEE379 pKa = 4.64SYY381 pKa = 10.96NEE383 pKa = 4.57LYY385 pKa = 10.74FEE387 pKa = 4.99QNGSDD392 pKa = 3.89NILVADD398 pKa = 3.71QRR400 pKa = 11.84GTGNYY405 pKa = 7.6ATGTSNGDD413 pKa = 3.48FNKK416 pKa = 9.82VEE418 pKa = 4.51LDD420 pKa = 3.23QSGAGNQAYY429 pKa = 10.32VNQLYY434 pKa = 10.65GDD436 pKa = 4.31HH437 pKa = 6.74NLASVTQTDD446 pKa = 4.57GVNLAYY452 pKa = 8.92ITQGGTANQATVSQAGVNMNANVQQFGTGNQSTITQQQ489 pKa = 2.8
MM1 pKa = 7.37FRR3 pKa = 11.84LTPISAALLLLAAGQAMADD22 pKa = 3.78DD23 pKa = 4.36SSANQVQNGEE33 pKa = 4.05QNIANVSQKK42 pKa = 9.46QASWSTANQTQTGTGNDD59 pKa = 3.13AMAVQQEE66 pKa = 4.58STGSIVQSLSGDD78 pKa = 3.15HH79 pKa = 5.93NAAYY83 pKa = 10.34GEE85 pKa = 4.11QNYY88 pKa = 10.38GEE90 pKa = 4.5INSSINQQGNGDD102 pKa = 3.72WNDD105 pKa = 3.4NFASQYY111 pKa = 10.45YY112 pKa = 10.56GSDD115 pKa = 3.26NQALQIQDD123 pKa = 3.37GSRR126 pKa = 11.84SFSMVYY132 pKa = 9.68QEE134 pKa = 4.66GQVGSQGTTQQSGGNHH150 pKa = 5.71EE151 pKa = 4.95ANIEE155 pKa = 4.01QIGFGEE161 pKa = 4.55GNRR164 pKa = 11.84ALIIQSGDD172 pKa = 3.17ANRR175 pKa = 11.84GSIKK179 pKa = 10.53QSDD182 pKa = 4.35QISGHH187 pKa = 4.93QEE189 pKa = 3.71VYY191 pKa = 10.25QQGSSNTANGWQNLGLGGTVLFNQAGDD218 pKa = 3.54NNYY221 pKa = 10.7AEE223 pKa = 4.67VWQDD227 pKa = 3.45SQDD230 pKa = 3.33SSQASIAQFGSTNEE244 pKa = 3.86AVIDD248 pKa = 3.84QSFGMAMDD256 pKa = 4.91PYY258 pKa = 11.13GVGNVASINQSGDD271 pKa = 3.17RR272 pKa = 11.84NAAWADD278 pKa = 3.47QYY280 pKa = 11.4EE281 pKa = 4.83SKK283 pKa = 11.02GSNTALYY290 pKa = 10.74QNGTDD295 pKa = 4.02NLQLTYY301 pKa = 9.66QTGSGHH307 pKa = 5.71SLVVTSNGTGNKK319 pKa = 9.52VYY321 pKa = 10.47MSDD324 pKa = 2.79WQGARR329 pKa = 11.84RR330 pKa = 11.84GGQSGQGNNAAIAQNGGGNVAQVKK354 pKa = 9.1QQDD357 pKa = 4.04TGNKK361 pKa = 7.52TQLTQNGTNNDD372 pKa = 3.8LNVSQAEE379 pKa = 4.64SYY381 pKa = 10.96NEE383 pKa = 4.57LYY385 pKa = 10.74FEE387 pKa = 4.99QNGSDD392 pKa = 3.89NILVADD398 pKa = 3.71QRR400 pKa = 11.84GTGNYY405 pKa = 7.6ATGTSNGDD413 pKa = 3.48FNKK416 pKa = 9.82VEE418 pKa = 4.51LDD420 pKa = 3.23QSGAGNQAYY429 pKa = 10.32VNQLYY434 pKa = 10.65GDD436 pKa = 4.31HH437 pKa = 6.74NLASVTQTDD446 pKa = 4.57GVNLAYY452 pKa = 8.92ITQGGTANQATVSQAGVNMNANVQQFGTGNQSTITQQQ489 pKa = 2.8
Molecular weight: 51.49 kDa
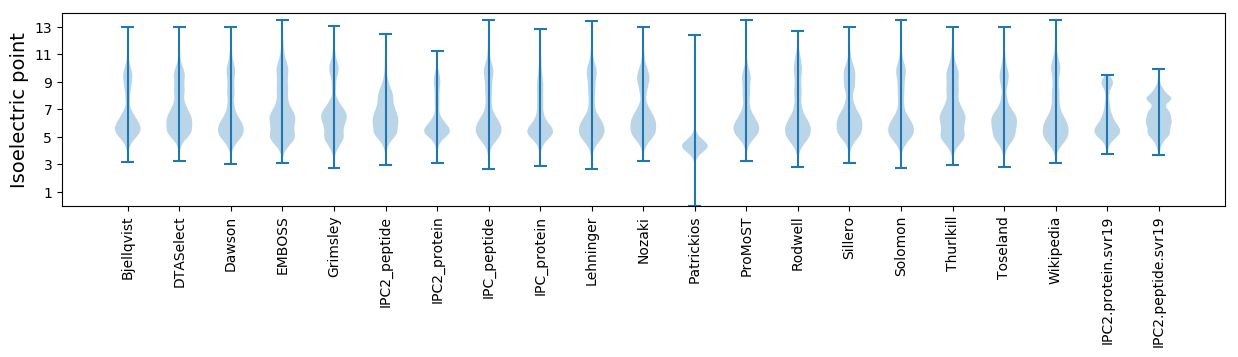
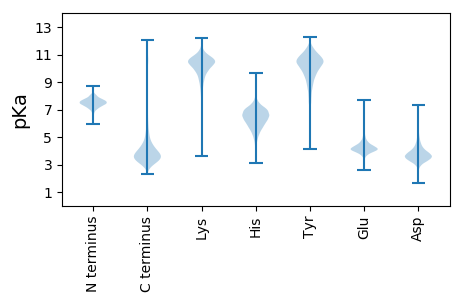
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024HQV8|A0A024HQV8_PSEKB Exodeoxyribonuclease III OS=Pseudomonas knackmussii (strain DSM 6978 / LMG 23759 / B13) OX=1301098 GN=crc PE=3 SV=1
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
MM1 pKa = 7.45KK2 pKa = 9.56RR3 pKa = 11.84TFQPSTLKK11 pKa = 10.31RR12 pKa = 11.84ARR14 pKa = 11.84VHH16 pKa = 6.26GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.29NGRR28 pKa = 11.84QVLSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.83GRR39 pKa = 11.84KK40 pKa = 8.88RR41 pKa = 11.84LTVV44 pKa = 3.11
Molecular weight: 5.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1780795 |
29 |
3787 |
317.1 |
34.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.552 ± 0.042 | 1.022 ± 0.011 |
5.351 ± 0.025 | 5.954 ± 0.036 |
3.573 ± 0.019 | 8.226 ± 0.031 |
2.269 ± 0.016 | 4.356 ± 0.023 |
3.196 ± 0.029 | 12.104 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.218 ± 0.014 | 2.77 ± 0.021 |
5.034 ± 0.026 | 4.429 ± 0.026 |
6.985 ± 0.034 | 5.557 ± 0.025 |
4.435 ± 0.03 | 6.989 ± 0.028 |
1.451 ± 0.013 | 2.528 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
