
Blackberry chlorotic ringspot virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Bromoviridae; Ilarvirus
Average proteome isoelectric point is 6.62
Get precalculated fractions of proteins
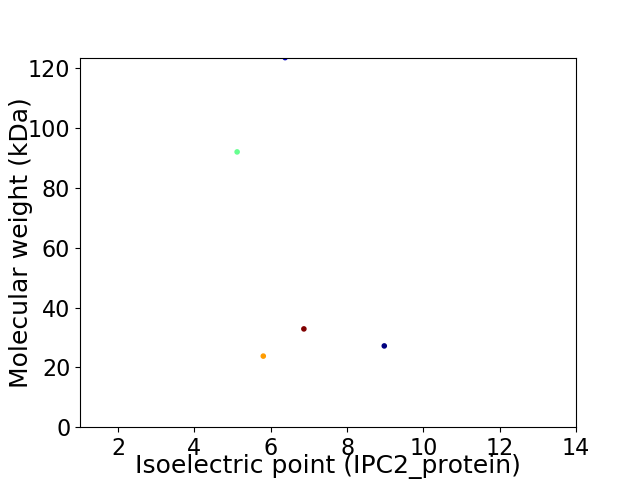
Virtual 2D-PAGE plot for 5 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q2QF35|Q2QF35_9BROM ATP-dependent helicase OS=Blackberry chlorotic ringspot virus OX=339420 PE=2 SV=1
MM1 pKa = 8.11DD2 pKa = 3.91YY3 pKa = 11.08VKK5 pKa = 10.83EE6 pKa = 3.89NLIVYY11 pKa = 7.5SLRR14 pKa = 11.84SRR16 pKa = 11.84LEE18 pKa = 4.16FIQSFGVEE26 pKa = 3.42PDD28 pKa = 3.39MYY30 pKa = 10.74KK31 pKa = 10.34EE32 pKa = 4.13WVSMFLFKK40 pKa = 10.75FIVEE44 pKa = 4.27HH45 pKa = 4.93TAKK48 pKa = 10.4FDD50 pKa = 3.67FATIDD55 pKa = 3.63CTFGMIVARR64 pKa = 11.84LIPGYY69 pKa = 10.58SDD71 pKa = 5.81LDD73 pKa = 4.35DD74 pKa = 4.53EE75 pKa = 5.62DD76 pKa = 5.39EE77 pKa = 4.18EE78 pKa = 4.41HH79 pKa = 6.85SPRR82 pKa = 11.84EE83 pKa = 3.52IDD85 pKa = 4.78SFYY88 pKa = 11.21LPYY91 pKa = 10.73DD92 pKa = 4.33DD93 pKa = 6.51LDD95 pKa = 3.56VDD97 pKa = 4.14YY98 pKa = 8.4TTIDD102 pKa = 3.37YY103 pKa = 10.85RR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.64DD107 pKa = 3.82VEE109 pKa = 4.39VPTNEE114 pKa = 3.85VLNLVSNASYY124 pKa = 10.9VPEE127 pKa = 4.34GVSWGDD133 pKa = 3.3VTDD136 pKa = 4.73SSFEE140 pKa = 3.9DD141 pKa = 3.41HH142 pKa = 7.15LEE144 pKa = 4.45EE145 pKa = 4.5IQEE148 pKa = 4.21LPTFNVDD155 pKa = 3.47EE156 pKa = 4.58PVVSSSEE163 pKa = 3.89AGLPDD168 pKa = 5.52DD169 pKa = 5.92DD170 pKa = 4.87GTIQDD175 pKa = 4.53VVWVDD180 pKa = 3.28QSDD183 pKa = 4.55PIVSDD188 pKa = 3.71VVADD192 pKa = 3.83ADD194 pKa = 3.97LRR196 pKa = 11.84NCGFVDD202 pKa = 3.81YY203 pKa = 10.84QSGSIKK209 pKa = 9.22TSKK212 pKa = 9.41WKK214 pKa = 10.21PKK216 pKa = 9.81VIQVRR221 pKa = 11.84PDD223 pKa = 3.12PSIIQNAVDD232 pKa = 4.22EE233 pKa = 4.74IFPNHH238 pKa = 6.04HH239 pKa = 6.65HH240 pKa = 6.8VDD242 pKa = 3.39DD243 pKa = 4.89KK244 pKa = 11.42FFQEE248 pKa = 4.07WVEE251 pKa = 3.92THH253 pKa = 7.34DD254 pKa = 4.63IDD256 pKa = 6.25LEE258 pKa = 4.44VTNCDD263 pKa = 3.15LDD265 pKa = 3.52MSVFNDD271 pKa = 3.25WTKK274 pKa = 11.19GSDD277 pKa = 3.62SRR279 pKa = 11.84LMPAFSVGGLSHH291 pKa = 6.8TVPTQRR297 pKa = 11.84EE298 pKa = 3.73TLLAVKK304 pKa = 9.36KK305 pKa = 10.66RR306 pKa = 11.84NMNVPEE312 pKa = 4.35LQSQFDD318 pKa = 4.09HH319 pKa = 6.84NDD321 pKa = 2.97VLNRR325 pKa = 11.84CVNRR329 pKa = 11.84FLTHH333 pKa = 6.03VVDD336 pKa = 3.81KK337 pKa = 10.85TRR339 pKa = 11.84LSKK342 pKa = 10.82LMPISGEE349 pKa = 3.87EE350 pKa = 3.55VHH352 pKa = 6.39YY353 pKa = 10.45FNQYY357 pKa = 9.67VEE359 pKa = 4.57NKK361 pKa = 9.71NPPLSEE367 pKa = 4.12YY368 pKa = 10.28KK369 pKa = 10.94GPIPLVALDD378 pKa = 5.02KK379 pKa = 10.39YY380 pKa = 9.66MHH382 pKa = 6.57MIKK385 pKa = 7.75TTLKK389 pKa = 10.13PVEE392 pKa = 4.31EE393 pKa = 5.13DD394 pKa = 3.26SLHH397 pKa = 5.79VEE399 pKa = 4.15RR400 pKa = 11.84PIPATITYY408 pKa = 8.34HH409 pKa = 5.89KK410 pKa = 9.69KK411 pKa = 9.51GVVMMTSPYY420 pKa = 9.61FLCAMVRR427 pKa = 11.84LLYY430 pKa = 10.08VLKK433 pKa = 10.74SKK435 pKa = 10.49FVVPTGKK442 pKa = 9.7YY443 pKa = 8.95HH444 pKa = 7.2QIFHH448 pKa = 6.94MDD450 pKa = 3.56PEE452 pKa = 4.24RR453 pKa = 11.84LEE455 pKa = 3.78VSNFFKK461 pKa = 10.9EE462 pKa = 3.6IDD464 pKa = 3.48FSKK467 pKa = 10.51FDD469 pKa = 3.37KK470 pKa = 11.04SQGRR474 pKa = 11.84LHH476 pKa = 7.1HH477 pKa = 7.07DD478 pKa = 3.4VQFKK482 pKa = 10.59LFLMLGIPEE491 pKa = 4.6HH492 pKa = 6.74FVTSWFNAHH501 pKa = 6.13EE502 pKa = 4.31LSHH505 pKa = 8.05IRR507 pKa = 11.84DD508 pKa = 3.65RR509 pKa = 11.84DD510 pKa = 3.8CGVGFSVDD518 pKa = 3.47YY519 pKa = 10.5QRR521 pKa = 11.84RR522 pKa = 11.84TGDD525 pKa = 2.74ACTYY529 pKa = 10.46LGNTLVTLSVLSYY542 pKa = 11.16VYY544 pKa = 10.73DD545 pKa = 4.08LSNPNISFVAASGDD559 pKa = 3.52DD560 pKa = 3.7SLIGSLQPLPRR571 pKa = 11.84DD572 pKa = 3.73KK573 pKa = 11.36EE574 pKa = 4.23DD575 pKa = 3.51LCVSLFNFEE584 pKa = 4.08TKK586 pKa = 10.2FPHH589 pKa = 5.6NQPFICSKK597 pKa = 9.9FLLVVEE603 pKa = 5.11CDD605 pKa = 3.75DD606 pKa = 3.73GTKK609 pKa = 10.35EE610 pKa = 3.99VLAVPNPLKK619 pKa = 10.61LLQKK623 pKa = 10.43LGPKK627 pKa = 8.78TMQVTMIDD635 pKa = 3.82DD636 pKa = 4.9YY637 pKa = 11.5YY638 pKa = 11.61QSLCDD643 pKa = 3.68ILWVFEE649 pKa = 4.4DD650 pKa = 3.94ADD652 pKa = 3.61VCRR655 pKa = 11.84RR656 pKa = 11.84VAEE659 pKa = 4.06LAEE662 pKa = 3.93FRR664 pKa = 11.84AFKK667 pKa = 10.53GRR669 pKa = 11.84KK670 pKa = 8.3KK671 pKa = 10.84CLFLEE676 pKa = 4.48SALLSLPSLVANRR689 pKa = 11.84LKK691 pKa = 10.7FLRR694 pKa = 11.84RR695 pKa = 11.84TINLEE700 pKa = 3.69SSKK703 pKa = 11.33ACIRR707 pKa = 11.84NDD709 pKa = 3.17VYY711 pKa = 11.84SNLVDD716 pKa = 3.98ALEE719 pKa = 4.28PMCARR724 pKa = 11.84RR725 pKa = 11.84LDD727 pKa = 3.71ASPRR731 pKa = 11.84RR732 pKa = 11.84SEE734 pKa = 3.68ICSGNEE740 pKa = 3.34KK741 pKa = 10.16VRR743 pKa = 11.84IGSRR747 pKa = 11.84NSRR750 pKa = 11.84CGSWRR755 pKa = 11.84KK756 pKa = 8.97EE757 pKa = 3.41YY758 pKa = 10.51RR759 pKa = 11.84AGDD762 pKa = 3.75KK763 pKa = 10.69IEE765 pKa = 4.01TPRR768 pKa = 11.84ICEE771 pKa = 4.09GVHH774 pKa = 5.42GRR776 pKa = 11.84GKK778 pKa = 10.33RR779 pKa = 11.84EE780 pKa = 3.66PSANPQRR787 pKa = 11.84ISFRR791 pKa = 11.84GKK793 pKa = 8.82EE794 pKa = 4.02LTWKK798 pKa = 10.37GGNKK802 pKa = 9.85LPP804 pKa = 4.75
MM1 pKa = 8.11DD2 pKa = 3.91YY3 pKa = 11.08VKK5 pKa = 10.83EE6 pKa = 3.89NLIVYY11 pKa = 7.5SLRR14 pKa = 11.84SRR16 pKa = 11.84LEE18 pKa = 4.16FIQSFGVEE26 pKa = 3.42PDD28 pKa = 3.39MYY30 pKa = 10.74KK31 pKa = 10.34EE32 pKa = 4.13WVSMFLFKK40 pKa = 10.75FIVEE44 pKa = 4.27HH45 pKa = 4.93TAKK48 pKa = 10.4FDD50 pKa = 3.67FATIDD55 pKa = 3.63CTFGMIVARR64 pKa = 11.84LIPGYY69 pKa = 10.58SDD71 pKa = 5.81LDD73 pKa = 4.35DD74 pKa = 4.53EE75 pKa = 5.62DD76 pKa = 5.39EE77 pKa = 4.18EE78 pKa = 4.41HH79 pKa = 6.85SPRR82 pKa = 11.84EE83 pKa = 3.52IDD85 pKa = 4.78SFYY88 pKa = 11.21LPYY91 pKa = 10.73DD92 pKa = 4.33DD93 pKa = 6.51LDD95 pKa = 3.56VDD97 pKa = 4.14YY98 pKa = 8.4TTIDD102 pKa = 3.37YY103 pKa = 10.85RR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.64DD107 pKa = 3.82VEE109 pKa = 4.39VPTNEE114 pKa = 3.85VLNLVSNASYY124 pKa = 10.9VPEE127 pKa = 4.34GVSWGDD133 pKa = 3.3VTDD136 pKa = 4.73SSFEE140 pKa = 3.9DD141 pKa = 3.41HH142 pKa = 7.15LEE144 pKa = 4.45EE145 pKa = 4.5IQEE148 pKa = 4.21LPTFNVDD155 pKa = 3.47EE156 pKa = 4.58PVVSSSEE163 pKa = 3.89AGLPDD168 pKa = 5.52DD169 pKa = 5.92DD170 pKa = 4.87GTIQDD175 pKa = 4.53VVWVDD180 pKa = 3.28QSDD183 pKa = 4.55PIVSDD188 pKa = 3.71VVADD192 pKa = 3.83ADD194 pKa = 3.97LRR196 pKa = 11.84NCGFVDD202 pKa = 3.81YY203 pKa = 10.84QSGSIKK209 pKa = 9.22TSKK212 pKa = 9.41WKK214 pKa = 10.21PKK216 pKa = 9.81VIQVRR221 pKa = 11.84PDD223 pKa = 3.12PSIIQNAVDD232 pKa = 4.22EE233 pKa = 4.74IFPNHH238 pKa = 6.04HH239 pKa = 6.65HH240 pKa = 6.8VDD242 pKa = 3.39DD243 pKa = 4.89KK244 pKa = 11.42FFQEE248 pKa = 4.07WVEE251 pKa = 3.92THH253 pKa = 7.34DD254 pKa = 4.63IDD256 pKa = 6.25LEE258 pKa = 4.44VTNCDD263 pKa = 3.15LDD265 pKa = 3.52MSVFNDD271 pKa = 3.25WTKK274 pKa = 11.19GSDD277 pKa = 3.62SRR279 pKa = 11.84LMPAFSVGGLSHH291 pKa = 6.8TVPTQRR297 pKa = 11.84EE298 pKa = 3.73TLLAVKK304 pKa = 9.36KK305 pKa = 10.66RR306 pKa = 11.84NMNVPEE312 pKa = 4.35LQSQFDD318 pKa = 4.09HH319 pKa = 6.84NDD321 pKa = 2.97VLNRR325 pKa = 11.84CVNRR329 pKa = 11.84FLTHH333 pKa = 6.03VVDD336 pKa = 3.81KK337 pKa = 10.85TRR339 pKa = 11.84LSKK342 pKa = 10.82LMPISGEE349 pKa = 3.87EE350 pKa = 3.55VHH352 pKa = 6.39YY353 pKa = 10.45FNQYY357 pKa = 9.67VEE359 pKa = 4.57NKK361 pKa = 9.71NPPLSEE367 pKa = 4.12YY368 pKa = 10.28KK369 pKa = 10.94GPIPLVALDD378 pKa = 5.02KK379 pKa = 10.39YY380 pKa = 9.66MHH382 pKa = 6.57MIKK385 pKa = 7.75TTLKK389 pKa = 10.13PVEE392 pKa = 4.31EE393 pKa = 5.13DD394 pKa = 3.26SLHH397 pKa = 5.79VEE399 pKa = 4.15RR400 pKa = 11.84PIPATITYY408 pKa = 8.34HH409 pKa = 5.89KK410 pKa = 9.69KK411 pKa = 9.51GVVMMTSPYY420 pKa = 9.61FLCAMVRR427 pKa = 11.84LLYY430 pKa = 10.08VLKK433 pKa = 10.74SKK435 pKa = 10.49FVVPTGKK442 pKa = 9.7YY443 pKa = 8.95HH444 pKa = 7.2QIFHH448 pKa = 6.94MDD450 pKa = 3.56PEE452 pKa = 4.24RR453 pKa = 11.84LEE455 pKa = 3.78VSNFFKK461 pKa = 10.9EE462 pKa = 3.6IDD464 pKa = 3.48FSKK467 pKa = 10.51FDD469 pKa = 3.37KK470 pKa = 11.04SQGRR474 pKa = 11.84LHH476 pKa = 7.1HH477 pKa = 7.07DD478 pKa = 3.4VQFKK482 pKa = 10.59LFLMLGIPEE491 pKa = 4.6HH492 pKa = 6.74FVTSWFNAHH501 pKa = 6.13EE502 pKa = 4.31LSHH505 pKa = 8.05IRR507 pKa = 11.84DD508 pKa = 3.65RR509 pKa = 11.84DD510 pKa = 3.8CGVGFSVDD518 pKa = 3.47YY519 pKa = 10.5QRR521 pKa = 11.84RR522 pKa = 11.84TGDD525 pKa = 2.74ACTYY529 pKa = 10.46LGNTLVTLSVLSYY542 pKa = 11.16VYY544 pKa = 10.73DD545 pKa = 4.08LSNPNISFVAASGDD559 pKa = 3.52DD560 pKa = 3.7SLIGSLQPLPRR571 pKa = 11.84DD572 pKa = 3.73KK573 pKa = 11.36EE574 pKa = 4.23DD575 pKa = 3.51LCVSLFNFEE584 pKa = 4.08TKK586 pKa = 10.2FPHH589 pKa = 5.6NQPFICSKK597 pKa = 9.9FLLVVEE603 pKa = 5.11CDD605 pKa = 3.75DD606 pKa = 3.73GTKK609 pKa = 10.35EE610 pKa = 3.99VLAVPNPLKK619 pKa = 10.61LLQKK623 pKa = 10.43LGPKK627 pKa = 8.78TMQVTMIDD635 pKa = 3.82DD636 pKa = 4.9YY637 pKa = 11.5YY638 pKa = 11.61QSLCDD643 pKa = 3.68ILWVFEE649 pKa = 4.4DD650 pKa = 3.94ADD652 pKa = 3.61VCRR655 pKa = 11.84RR656 pKa = 11.84VAEE659 pKa = 4.06LAEE662 pKa = 3.93FRR664 pKa = 11.84AFKK667 pKa = 10.53GRR669 pKa = 11.84KK670 pKa = 8.3KK671 pKa = 10.84CLFLEE676 pKa = 4.48SALLSLPSLVANRR689 pKa = 11.84LKK691 pKa = 10.7FLRR694 pKa = 11.84RR695 pKa = 11.84TINLEE700 pKa = 3.69SSKK703 pKa = 11.33ACIRR707 pKa = 11.84NDD709 pKa = 3.17VYY711 pKa = 11.84SNLVDD716 pKa = 3.98ALEE719 pKa = 4.28PMCARR724 pKa = 11.84RR725 pKa = 11.84LDD727 pKa = 3.71ASPRR731 pKa = 11.84RR732 pKa = 11.84SEE734 pKa = 3.68ICSGNEE740 pKa = 3.34KK741 pKa = 10.16VRR743 pKa = 11.84IGSRR747 pKa = 11.84NSRR750 pKa = 11.84CGSWRR755 pKa = 11.84KK756 pKa = 8.97EE757 pKa = 3.41YY758 pKa = 10.51RR759 pKa = 11.84AGDD762 pKa = 3.75KK763 pKa = 10.69IEE765 pKa = 4.01TPRR768 pKa = 11.84ICEE771 pKa = 4.09GVHH774 pKa = 5.42GRR776 pKa = 11.84GKK778 pKa = 10.33RR779 pKa = 11.84EE780 pKa = 3.66PSANPQRR787 pKa = 11.84ISFRR791 pKa = 11.84GKK793 pKa = 8.82EE794 pKa = 4.02LTWKK798 pKa = 10.37GGNKK802 pKa = 9.85LPP804 pKa = 4.75
Molecular weight: 92.06 kDa
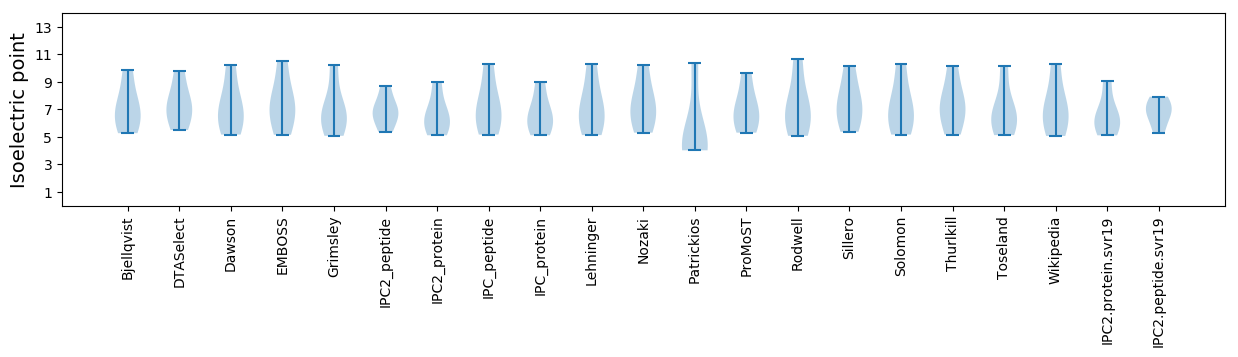
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q2QF33|Q2QF33_9BROM Movement protein OS=Blackberry chlorotic ringspot virus OX=339420 PE=2 SV=1
MM1 pKa = 7.13STLTKK6 pKa = 11.03NPDD9 pKa = 3.37QPSNAMSANRR19 pKa = 11.84GRR21 pKa = 11.84NGISNSNRR29 pKa = 11.84GCPNCFDD36 pKa = 4.56NLDD39 pKa = 3.87AAASNCTRR47 pKa = 11.84CNPAVSNRR55 pKa = 11.84QRR57 pKa = 11.84RR58 pKa = 11.84NARR61 pKa = 11.84RR62 pKa = 11.84AANFRR67 pKa = 11.84NQRR70 pKa = 11.84KK71 pKa = 9.03FDD73 pKa = 3.79GLRR76 pKa = 11.84ALQAPVPLPVVPVPQPATQRR96 pKa = 11.84NLRR99 pKa = 11.84LPNGQVWVTRR109 pKa = 11.84KK110 pKa = 7.97PTDD113 pKa = 2.9WAAKK117 pKa = 10.44VNDD120 pKa = 4.59ANDD123 pKa = 3.2AMLLKK128 pKa = 10.04TIFDD132 pKa = 5.55GITEE136 pKa = 4.7IKK138 pKa = 10.15PDD140 pKa = 3.43TKK142 pKa = 10.72VFRR145 pKa = 11.84VLIGFVAMSDD155 pKa = 3.62GTFGLVDD162 pKa = 4.29GVTGDD167 pKa = 4.01TVPEE171 pKa = 3.91LPIIGRR177 pKa = 11.84WSFQRR182 pKa = 11.84DD183 pKa = 3.66VYY185 pKa = 10.58RR186 pKa = 11.84SRR188 pKa = 11.84DD189 pKa = 3.31IGLDD193 pKa = 3.54GQPADD198 pKa = 3.75QLSEE202 pKa = 4.19KK203 pKa = 10.67AVVWCLNTNKK213 pKa = 10.09RR214 pKa = 11.84AEE216 pKa = 4.1KK217 pKa = 10.01RR218 pKa = 11.84VRR220 pKa = 11.84LADD223 pKa = 3.92FWVAIAKK230 pKa = 7.5PKK232 pKa = 10.38PLMPPPDD239 pKa = 4.16FLVEE243 pKa = 4.42DD244 pKa = 3.88NN245 pKa = 4.33
MM1 pKa = 7.13STLTKK6 pKa = 11.03NPDD9 pKa = 3.37QPSNAMSANRR19 pKa = 11.84GRR21 pKa = 11.84NGISNSNRR29 pKa = 11.84GCPNCFDD36 pKa = 4.56NLDD39 pKa = 3.87AAASNCTRR47 pKa = 11.84CNPAVSNRR55 pKa = 11.84QRR57 pKa = 11.84RR58 pKa = 11.84NARR61 pKa = 11.84RR62 pKa = 11.84AANFRR67 pKa = 11.84NQRR70 pKa = 11.84KK71 pKa = 9.03FDD73 pKa = 3.79GLRR76 pKa = 11.84ALQAPVPLPVVPVPQPATQRR96 pKa = 11.84NLRR99 pKa = 11.84LPNGQVWVTRR109 pKa = 11.84KK110 pKa = 7.97PTDD113 pKa = 2.9WAAKK117 pKa = 10.44VNDD120 pKa = 4.59ANDD123 pKa = 3.2AMLLKK128 pKa = 10.04TIFDD132 pKa = 5.55GITEE136 pKa = 4.7IKK138 pKa = 10.15PDD140 pKa = 3.43TKK142 pKa = 10.72VFRR145 pKa = 11.84VLIGFVAMSDD155 pKa = 3.62GTFGLVDD162 pKa = 4.29GVTGDD167 pKa = 4.01TVPEE171 pKa = 3.91LPIIGRR177 pKa = 11.84WSFQRR182 pKa = 11.84DD183 pKa = 3.66VYY185 pKa = 10.58RR186 pKa = 11.84SRR188 pKa = 11.84DD189 pKa = 3.31IGLDD193 pKa = 3.54GQPADD198 pKa = 3.75QLSEE202 pKa = 4.19KK203 pKa = 10.67AVVWCLNTNKK213 pKa = 10.09RR214 pKa = 11.84AEE216 pKa = 4.1KK217 pKa = 10.01RR218 pKa = 11.84VRR220 pKa = 11.84LADD223 pKa = 3.92FWVAIAKK230 pKa = 7.5PKK232 pKa = 10.38PLMPPPDD239 pKa = 4.16FLVEE243 pKa = 4.42DD244 pKa = 3.88NN245 pKa = 4.33
Molecular weight: 27.18 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2652 |
211 |
1093 |
530.4 |
59.9 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.41 ± 1.073 | 2.262 ± 0.182 |
7.315 ± 0.611 | 5.882 ± 0.664 |
4.299 ± 0.409 | 4.299 ± 0.258 |
2.376 ± 0.41 | 5.204 ± 0.343 |
6.184 ± 0.331 | 8.71 ± 0.279 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.64 ± 0.187 | 4.751 ± 0.579 |
5.166 ± 0.988 | 2.376 ± 0.3 |
6.109 ± 0.471 | 7.541 ± 0.439 |
5.807 ± 0.559 | 8.71 ± 0.174 |
1.131 ± 0.153 | 2.828 ± 0.579 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
