
Candidatus Brocadiaceae bacterium S225
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Candidatus Brocadiae; Candidatus Brocadiales; Candidatus Brocadiaceae; unclassified Candidatus Brocadiaceae
Average proteome isoelectric point is 6.72
Get precalculated fractions of proteins
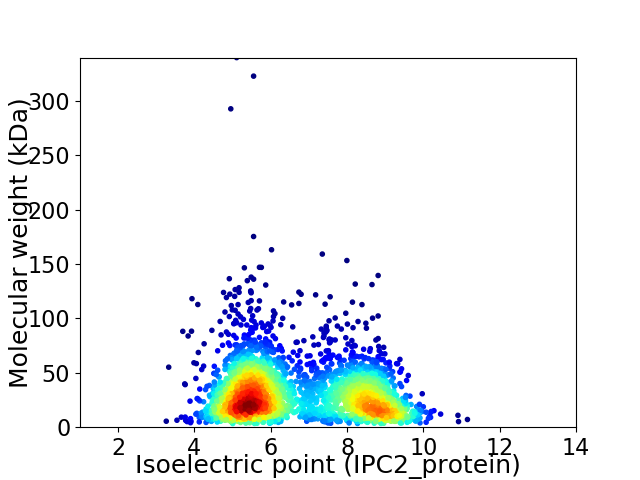
Virtual 2D-PAGE plot for 2768 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C6D5K1|A0A5C6D5K1_9BACT Cobalt-precorrin-2 C(20)-methyltransferase OS=Candidatus Brocadiaceae bacterium S225 OX=2528033 GN=cbiL PE=3 SV=1
MM1 pKa = 7.97RR2 pKa = 11.84IKK4 pKa = 10.96NFIIAVLVAVVVGVSGGLATAQITNFSTDD33 pKa = 2.01VDD35 pKa = 3.93TAINRR40 pKa = 11.84GLTFQDD46 pKa = 2.97SSGSYY51 pKa = 9.06TGSAGDD57 pKa = 3.67ATGLALLSLLEE68 pKa = 4.27KK69 pKa = 10.4RR70 pKa = 11.84VSADD74 pKa = 3.13QNAAPQGYY82 pKa = 9.7VNANAADD89 pKa = 3.84QNRR92 pKa = 11.84MNSAVDD98 pKa = 4.6FIIDD102 pKa = 4.05EE103 pKa = 4.06NRR105 pKa = 11.84EE106 pKa = 3.99DD107 pKa = 3.87NGCGGEE113 pKa = 4.71DD114 pKa = 4.19EE115 pKa = 5.31NCAPCNTEE123 pKa = 3.65WADD126 pKa = 3.2GDD128 pKa = 3.92AADD131 pKa = 3.83WQNLSFEE138 pKa = 4.68PFRR141 pKa = 11.84DD142 pKa = 3.18WSGRR146 pKa = 11.84GQGPNMLGRR155 pKa = 11.84PAVLHH160 pKa = 6.58LIEE163 pKa = 4.51EE164 pKa = 4.91DD165 pKa = 3.12IYY167 pKa = 11.5LDD169 pKa = 4.45VEE171 pKa = 4.18FTLWDD176 pKa = 3.37TCGFGSEE183 pKa = 4.77GEE185 pKa = 4.4GEE187 pKa = 4.43GEE189 pKa = 4.35GEE191 pKa = 4.4GEE193 pKa = 4.39GEE195 pKa = 4.47GEE197 pKa = 4.37GGSGSAVQVGGCSPFPGFQYY217 pKa = 10.67LRR219 pKa = 11.84STEE222 pKa = 4.15GVPGEE227 pKa = 4.28KK228 pKa = 9.92VWTGGQITFTKK239 pKa = 10.19PCGADD244 pKa = 3.0PDD246 pKa = 4.27VPANGDD252 pKa = 3.61GITAFTRR259 pKa = 11.84LTRR262 pKa = 11.84GDD264 pKa = 3.75CGSIYY269 pKa = 10.78NSVCEE274 pKa = 3.86EE275 pKa = 4.36DD276 pKa = 4.5FFNSAPCPGGPGGLPFYY293 pKa = 10.5AYY295 pKa = 10.3RR296 pKa = 11.84DD297 pKa = 4.17GQDD300 pKa = 2.88MMALSVYY307 pKa = 9.87RR308 pKa = 11.84RR309 pKa = 11.84TGGPIDD315 pKa = 3.95VLPALLAIFDD325 pKa = 3.99RR326 pKa = 11.84TIAAQAPDD334 pKa = 5.44GYY336 pKa = 10.1WCYY339 pKa = 11.05CDD341 pKa = 5.25GGCSDD346 pKa = 3.96SSTTQFVIAGLAAIRR361 pKa = 11.84AVLIDD366 pKa = 3.91EE367 pKa = 4.62NDD369 pKa = 3.59FARR372 pKa = 11.84IAQLDD377 pKa = 3.86AATALCRR384 pKa = 11.84AAYY387 pKa = 10.13ASIPAQNPVLTPGEE401 pKa = 4.28LGHH404 pKa = 6.76GYY406 pKa = 10.14NRR408 pKa = 11.84GNQNSLQQTGSGTWIQLAGGADD430 pKa = 3.75LNDD433 pKa = 4.35AGVQAYY439 pKa = 9.19LRR441 pKa = 11.84WLYY444 pKa = 11.04NRR446 pKa = 11.84YY447 pKa = 9.21NDD449 pKa = 4.72RR450 pKa = 11.84SILGAGGGWSGSYY463 pKa = 10.24FYY465 pKa = 10.51YY466 pKa = 10.4LWSFAKK472 pKa = 10.62ALTFLEE478 pKa = 4.75DD479 pKa = 3.62SGITANAGNLAVADD493 pKa = 4.97LGTLDD498 pKa = 4.19PAAVPAFGGRR508 pKa = 11.84VLHH511 pKa = 7.06RR512 pKa = 11.84DD513 pKa = 3.3PTTVPRR519 pKa = 11.84PAVFGPDD526 pKa = 2.96PGAYY530 pKa = 9.8YY531 pKa = 10.31DD532 pKa = 5.48DD533 pKa = 5.1PNEE536 pKa = 3.96PPRR539 pKa = 11.84WYY541 pKa = 10.66FDD543 pKa = 3.31FAYY546 pKa = 10.7SLLLLQDD553 pKa = 3.73ASGQFLNPAGHH564 pKa = 6.5GNWDD568 pKa = 3.21NAARR572 pKa = 11.84QAYY575 pKa = 9.39SLLVLVRR582 pKa = 11.84SVGGGCLDD590 pKa = 3.38TDD592 pKa = 4.13GDD594 pKa = 4.29QVCDD598 pKa = 4.72SEE600 pKa = 5.31DD601 pKa = 3.53NCPADD606 pKa = 4.08ANPGQEE612 pKa = 5.42DD613 pKa = 4.12GDD615 pKa = 4.13SDD617 pKa = 5.05DD618 pKa = 5.44VGDD621 pKa = 4.6VCDD624 pKa = 3.95NCINTPNPGQEE635 pKa = 4.95DD636 pKa = 4.1GDD638 pKa = 4.05SDD640 pKa = 4.63GVGDD644 pKa = 5.06ACDD647 pKa = 3.87NCPDD651 pKa = 4.63DD652 pKa = 5.86ANPNQDD658 pKa = 4.8DD659 pKa = 4.07GDD661 pKa = 4.3GDD663 pKa = 4.41GVGDD667 pKa = 3.98VCDD670 pKa = 3.92NCLDD674 pKa = 4.07APNPNQADD682 pKa = 3.62GDD684 pKa = 4.04NDD686 pKa = 4.19GLGDD690 pKa = 4.29ACDD693 pKa = 3.98NCPDD697 pKa = 4.81DD698 pKa = 5.76ANPNQGDD705 pKa = 3.43VDD707 pKa = 3.99GDD709 pKa = 4.29GVGDD713 pKa = 3.75VCDD716 pKa = 3.89NCIDD720 pKa = 3.93TPNPGQEE727 pKa = 4.57DD728 pKa = 3.71VDD730 pKa = 4.05GDD732 pKa = 4.52GVGDD736 pKa = 4.07ACDD739 pKa = 3.76TPQIPCVEE747 pKa = 4.54DD748 pKa = 3.0LTARR752 pKa = 11.84AKK754 pKa = 9.55TGQVTLVWTHH764 pKa = 5.94VGADD768 pKa = 3.42SYY770 pKa = 11.86NVYY773 pKa = 10.72SGLNPGGPYY782 pKa = 10.76GLIGNTTSTYY792 pKa = 9.45STYY795 pKa = 10.63TDD797 pKa = 2.97SGLTNGITVYY807 pKa = 10.79YY808 pKa = 9.74IVRR811 pKa = 11.84PVLGGQEE818 pKa = 4.15LCPVTGNEE826 pKa = 4.24ASEE829 pKa = 4.57TPQEE833 pKa = 3.73RR834 pKa = 11.84RR835 pKa = 11.84RR836 pKa = 11.84RR837 pKa = 11.84EE838 pKa = 3.62RR839 pKa = 3.48
MM1 pKa = 7.97RR2 pKa = 11.84IKK4 pKa = 10.96NFIIAVLVAVVVGVSGGLATAQITNFSTDD33 pKa = 2.01VDD35 pKa = 3.93TAINRR40 pKa = 11.84GLTFQDD46 pKa = 2.97SSGSYY51 pKa = 9.06TGSAGDD57 pKa = 3.67ATGLALLSLLEE68 pKa = 4.27KK69 pKa = 10.4RR70 pKa = 11.84VSADD74 pKa = 3.13QNAAPQGYY82 pKa = 9.7VNANAADD89 pKa = 3.84QNRR92 pKa = 11.84MNSAVDD98 pKa = 4.6FIIDD102 pKa = 4.05EE103 pKa = 4.06NRR105 pKa = 11.84EE106 pKa = 3.99DD107 pKa = 3.87NGCGGEE113 pKa = 4.71DD114 pKa = 4.19EE115 pKa = 5.31NCAPCNTEE123 pKa = 3.65WADD126 pKa = 3.2GDD128 pKa = 3.92AADD131 pKa = 3.83WQNLSFEE138 pKa = 4.68PFRR141 pKa = 11.84DD142 pKa = 3.18WSGRR146 pKa = 11.84GQGPNMLGRR155 pKa = 11.84PAVLHH160 pKa = 6.58LIEE163 pKa = 4.51EE164 pKa = 4.91DD165 pKa = 3.12IYY167 pKa = 11.5LDD169 pKa = 4.45VEE171 pKa = 4.18FTLWDD176 pKa = 3.37TCGFGSEE183 pKa = 4.77GEE185 pKa = 4.4GEE187 pKa = 4.43GEE189 pKa = 4.35GEE191 pKa = 4.4GEE193 pKa = 4.39GEE195 pKa = 4.47GEE197 pKa = 4.37GGSGSAVQVGGCSPFPGFQYY217 pKa = 10.67LRR219 pKa = 11.84STEE222 pKa = 4.15GVPGEE227 pKa = 4.28KK228 pKa = 9.92VWTGGQITFTKK239 pKa = 10.19PCGADD244 pKa = 3.0PDD246 pKa = 4.27VPANGDD252 pKa = 3.61GITAFTRR259 pKa = 11.84LTRR262 pKa = 11.84GDD264 pKa = 3.75CGSIYY269 pKa = 10.78NSVCEE274 pKa = 3.86EE275 pKa = 4.36DD276 pKa = 4.5FFNSAPCPGGPGGLPFYY293 pKa = 10.5AYY295 pKa = 10.3RR296 pKa = 11.84DD297 pKa = 4.17GQDD300 pKa = 2.88MMALSVYY307 pKa = 9.87RR308 pKa = 11.84RR309 pKa = 11.84TGGPIDD315 pKa = 3.95VLPALLAIFDD325 pKa = 3.99RR326 pKa = 11.84TIAAQAPDD334 pKa = 5.44GYY336 pKa = 10.1WCYY339 pKa = 11.05CDD341 pKa = 5.25GGCSDD346 pKa = 3.96SSTTQFVIAGLAAIRR361 pKa = 11.84AVLIDD366 pKa = 3.91EE367 pKa = 4.62NDD369 pKa = 3.59FARR372 pKa = 11.84IAQLDD377 pKa = 3.86AATALCRR384 pKa = 11.84AAYY387 pKa = 10.13ASIPAQNPVLTPGEE401 pKa = 4.28LGHH404 pKa = 6.76GYY406 pKa = 10.14NRR408 pKa = 11.84GNQNSLQQTGSGTWIQLAGGADD430 pKa = 3.75LNDD433 pKa = 4.35AGVQAYY439 pKa = 9.19LRR441 pKa = 11.84WLYY444 pKa = 11.04NRR446 pKa = 11.84YY447 pKa = 9.21NDD449 pKa = 4.72RR450 pKa = 11.84SILGAGGGWSGSYY463 pKa = 10.24FYY465 pKa = 10.51YY466 pKa = 10.4LWSFAKK472 pKa = 10.62ALTFLEE478 pKa = 4.75DD479 pKa = 3.62SGITANAGNLAVADD493 pKa = 4.97LGTLDD498 pKa = 4.19PAAVPAFGGRR508 pKa = 11.84VLHH511 pKa = 7.06RR512 pKa = 11.84DD513 pKa = 3.3PTTVPRR519 pKa = 11.84PAVFGPDD526 pKa = 2.96PGAYY530 pKa = 9.8YY531 pKa = 10.31DD532 pKa = 5.48DD533 pKa = 5.1PNEE536 pKa = 3.96PPRR539 pKa = 11.84WYY541 pKa = 10.66FDD543 pKa = 3.31FAYY546 pKa = 10.7SLLLLQDD553 pKa = 3.73ASGQFLNPAGHH564 pKa = 6.5GNWDD568 pKa = 3.21NAARR572 pKa = 11.84QAYY575 pKa = 9.39SLLVLVRR582 pKa = 11.84SVGGGCLDD590 pKa = 3.38TDD592 pKa = 4.13GDD594 pKa = 4.29QVCDD598 pKa = 4.72SEE600 pKa = 5.31DD601 pKa = 3.53NCPADD606 pKa = 4.08ANPGQEE612 pKa = 5.42DD613 pKa = 4.12GDD615 pKa = 4.13SDD617 pKa = 5.05DD618 pKa = 5.44VGDD621 pKa = 4.6VCDD624 pKa = 3.95NCINTPNPGQEE635 pKa = 4.95DD636 pKa = 4.1GDD638 pKa = 4.05SDD640 pKa = 4.63GVGDD644 pKa = 5.06ACDD647 pKa = 3.87NCPDD651 pKa = 4.63DD652 pKa = 5.86ANPNQDD658 pKa = 4.8DD659 pKa = 4.07GDD661 pKa = 4.3GDD663 pKa = 4.41GVGDD667 pKa = 3.98VCDD670 pKa = 3.92NCLDD674 pKa = 4.07APNPNQADD682 pKa = 3.62GDD684 pKa = 4.04NDD686 pKa = 4.19GLGDD690 pKa = 4.29ACDD693 pKa = 3.98NCPDD697 pKa = 4.81DD698 pKa = 5.76ANPNQGDD705 pKa = 3.43VDD707 pKa = 3.99GDD709 pKa = 4.29GVGDD713 pKa = 3.75VCDD716 pKa = 3.89NCIDD720 pKa = 3.93TPNPGQEE727 pKa = 4.57DD728 pKa = 3.71VDD730 pKa = 4.05GDD732 pKa = 4.52GVGDD736 pKa = 4.07ACDD739 pKa = 3.76TPQIPCVEE747 pKa = 4.54DD748 pKa = 3.0LTARR752 pKa = 11.84AKK754 pKa = 9.55TGQVTLVWTHH764 pKa = 5.94VGADD768 pKa = 3.42SYY770 pKa = 11.86NVYY773 pKa = 10.72SGLNPGGPYY782 pKa = 10.76GLIGNTTSTYY792 pKa = 9.45STYY795 pKa = 10.63TDD797 pKa = 2.97SGLTNGITVYY807 pKa = 10.79YY808 pKa = 9.74IVRR811 pKa = 11.84PVLGGQEE818 pKa = 4.15LCPVTGNEE826 pKa = 4.24ASEE829 pKa = 4.57TPQEE833 pKa = 3.73RR834 pKa = 11.84RR835 pKa = 11.84RR836 pKa = 11.84RR837 pKa = 11.84EE838 pKa = 3.62RR839 pKa = 3.48
Molecular weight: 88.1 kDa
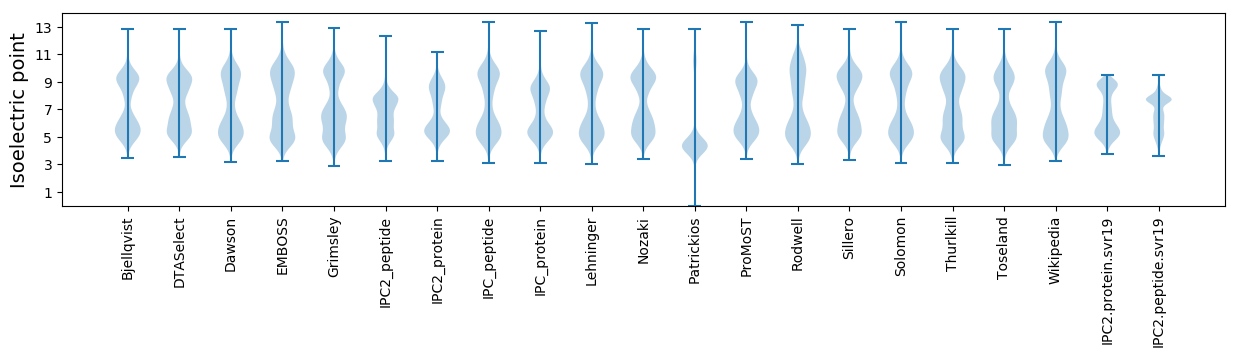
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C6DFI0|A0A5C6DFI0_9BACT Hydroxylamine reductase OS=Candidatus Brocadiaceae bacterium S225 OX=2528033 GN=hcp PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 10.57LKK5 pKa = 8.67THH7 pKa = 6.6KK8 pKa = 10.54GLTKK12 pKa = 10.15RR13 pKa = 11.84VRR15 pKa = 11.84VSSNGKK21 pKa = 9.2IKK23 pKa = 10.41RR24 pKa = 11.84KK25 pKa = 9.42KK26 pKa = 10.17AGAGHH31 pKa = 7.33LMSGKK36 pKa = 8.08TGSRR40 pKa = 11.84KK41 pKa = 8.51RR42 pKa = 11.84TLKK45 pKa = 9.78RR46 pKa = 11.84TSLVSKK52 pKa = 10.42GFTRR56 pKa = 11.84SVLRR60 pKa = 11.84ALGKK64 pKa = 10.33AA65 pKa = 3.52
MM1 pKa = 7.84PKK3 pKa = 10.57LKK5 pKa = 8.67THH7 pKa = 6.6KK8 pKa = 10.54GLTKK12 pKa = 10.15RR13 pKa = 11.84VRR15 pKa = 11.84VSSNGKK21 pKa = 9.2IKK23 pKa = 10.41RR24 pKa = 11.84KK25 pKa = 9.42KK26 pKa = 10.17AGAGHH31 pKa = 7.33LMSGKK36 pKa = 8.08TGSRR40 pKa = 11.84KK41 pKa = 8.51RR42 pKa = 11.84TLKK45 pKa = 9.78RR46 pKa = 11.84TSLVSKK52 pKa = 10.42GFTRR56 pKa = 11.84SVLRR60 pKa = 11.84ALGKK64 pKa = 10.33AA65 pKa = 3.52
Molecular weight: 7.09 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
835022 |
29 |
2953 |
301.7 |
33.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.381 ± 0.04 | 1.296 ± 0.023 |
5.639 ± 0.037 | 6.952 ± 0.045 |
4.509 ± 0.037 | 6.79 ± 0.044 |
1.982 ± 0.022 | 8.416 ± 0.049 |
7.591 ± 0.054 | 9.351 ± 0.051 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.464 ± 0.024 | 4.79 ± 0.035 |
3.584 ± 0.025 | 2.851 ± 0.023 |
4.637 ± 0.037 | 6.463 ± 0.035 |
5.288 ± 0.037 | 6.612 ± 0.039 |
1.002 ± 0.016 | 3.404 ± 0.03 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
