
Metallosphaera sedula (strain ATCC 51363 / DSM 5348 / JCM 9185 / NBRC 15509 / TH2)
Taxonomy: cellular organisms; Archaea; TACK group; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae; Metallosphaera; Metallosphaera sedula
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
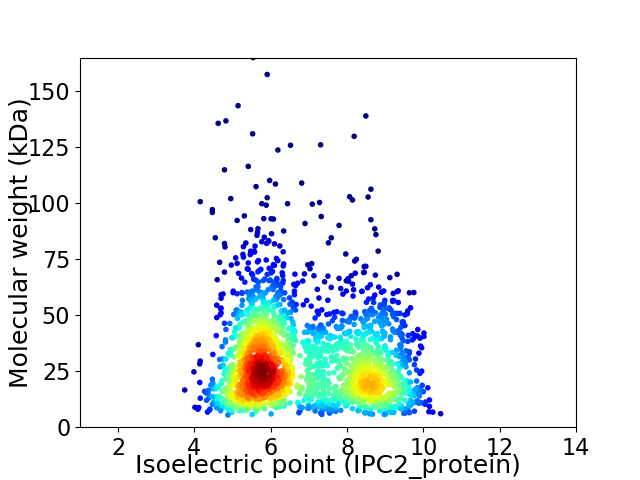
Virtual 2D-PAGE plot for 2256 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
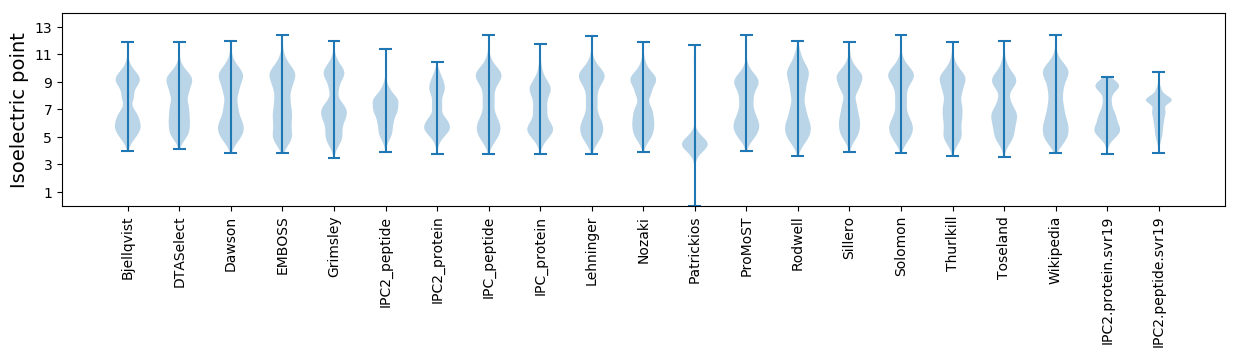
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A4YG22|A4YG22_METS5 2-oxoacid oxidoreductase (ferredoxin) OS=Metallosphaera sedula (strain ATCC 51363 / DSM 5348 / JCM 9185 / NBRC 15509 / TH2) OX=399549 GN=Msed_1213 PE=4 SV=1
MM1 pKa = 7.58SKK3 pKa = 10.02ILLILLGLVLASGAVVLGSNVNNVTSNIIHH33 pKa = 6.33QINPAVQGQQGTHH46 pKa = 6.93DD47 pKa = 4.02NSPDD51 pKa = 3.2NSTNSTNDD59 pKa = 3.17NTPDD63 pKa = 3.18NSTAIIHH70 pKa = 7.01DD71 pKa = 3.99NSPDD75 pKa = 3.18NSTNSTYY82 pKa = 11.07DD83 pKa = 3.41DD84 pKa = 3.8SHH86 pKa = 8.71HH87 pKa = 7.65DD88 pKa = 3.74DD89 pKa = 4.18STNSTYY95 pKa = 11.04SDD97 pKa = 4.54DD98 pKa = 3.75GHH100 pKa = 8.26HH101 pKa = 7.64DD102 pKa = 3.72DD103 pKa = 5.1SYY105 pKa = 12.11QNGTYY110 pKa = 10.2TNSTYY115 pKa = 11.11DD116 pKa = 4.19DD117 pKa = 3.8SHH119 pKa = 8.41HH120 pKa = 7.49DD121 pKa = 3.37NSTNSTYY128 pKa = 11.28DD129 pKa = 3.36NSPDD133 pKa = 3.2NSTNTNSTYY142 pKa = 11.11DD143 pKa = 3.48NSPDD147 pKa = 3.43QGSSGEE153 pKa = 4.11GDD155 pKa = 3.37SS156 pKa = 4.3
MM1 pKa = 7.58SKK3 pKa = 10.02ILLILLGLVLASGAVVLGSNVNNVTSNIIHH33 pKa = 6.33QINPAVQGQQGTHH46 pKa = 6.93DD47 pKa = 4.02NSPDD51 pKa = 3.2NSTNSTNDD59 pKa = 3.17NTPDD63 pKa = 3.18NSTAIIHH70 pKa = 7.01DD71 pKa = 3.99NSPDD75 pKa = 3.18NSTNSTYY82 pKa = 11.07DD83 pKa = 3.41DD84 pKa = 3.8SHH86 pKa = 8.71HH87 pKa = 7.65DD88 pKa = 3.74DD89 pKa = 4.18STNSTYY95 pKa = 11.04SDD97 pKa = 4.54DD98 pKa = 3.75GHH100 pKa = 8.26HH101 pKa = 7.64DD102 pKa = 3.72DD103 pKa = 5.1SYY105 pKa = 12.11QNGTYY110 pKa = 10.2TNSTYY115 pKa = 11.11DD116 pKa = 4.19DD117 pKa = 3.8SHH119 pKa = 8.41HH120 pKa = 7.49DD121 pKa = 3.37NSTNSTYY128 pKa = 11.28DD129 pKa = 3.36NSPDD133 pKa = 3.2NSTNTNSTYY142 pKa = 11.11DD143 pKa = 3.48NSPDD147 pKa = 3.43QGSSGEE153 pKa = 4.11GDD155 pKa = 3.37SS156 pKa = 4.3
Molecular weight: 16.61 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A4YIA4|A4YIA4_METS5 FeoA family protein OS=Metallosphaera sedula (strain ATCC 51363 / DSM 5348 / JCM 9185 / NBRC 15509 / TH2) OX=399549 GN=Msed_2016 PE=3 SV=1
MM1 pKa = 7.95RR2 pKa = 11.84LGKK5 pKa = 9.99ILGEE9 pKa = 3.81NRR11 pKa = 11.84SFRR14 pKa = 11.84FYY16 pKa = 9.67WASISLSHH24 pKa = 7.27ISGIMFNVISAYY36 pKa = 9.58ILLLVSPSFYY46 pKa = 10.73SLVVGVTMLVGALIRR61 pKa = 11.84LPSGYY66 pKa = 10.52LSDD69 pKa = 3.74SLDD72 pKa = 3.34RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.65ALMLTRR81 pKa = 11.84LSQAVLILLPILSLKK96 pKa = 8.45LTPFSYY102 pKa = 10.96VGFNLLTSLNGPLSAGLVQSVMEE125 pKa = 4.13KK126 pKa = 10.11RR127 pKa = 11.84QVLGGTSLNSLTISISSVVGGLVSLSYY154 pKa = 10.34PVLGITAFLSIASVMRR170 pKa = 11.84LFTVFFIGNVRR181 pKa = 11.84VKK183 pKa = 10.63ARR185 pKa = 11.84KK186 pKa = 9.51DD187 pKa = 3.48EE188 pKa = 4.46PKK190 pKa = 10.8GRR192 pKa = 11.84IPVSLIIYY200 pKa = 7.09PILVFSFLDD209 pKa = 3.95VIPVMMNSLVFVVSSVEE226 pKa = 3.64PSLALVSLLTTLGNGMGAILTGEE249 pKa = 4.73RR250 pKa = 11.84IRR252 pKa = 11.84EE253 pKa = 3.78NSIGIWARR261 pKa = 11.84TGSLLLGLVLVAISLSPLYY280 pKa = 10.24LVYY283 pKa = 10.89VLLVLRR289 pKa = 11.84GILSSIQSISIRR301 pKa = 11.84SDD303 pKa = 2.67LRR305 pKa = 11.84LRR307 pKa = 11.84VSNNLMGRR315 pKa = 11.84TWGLITTISSFVASVVALSYY335 pKa = 11.22SFIVSVTGTRR345 pKa = 11.84FPVLVLGLSLLTLGILMVYY364 pKa = 10.01RR365 pKa = 11.84GPSNRR370 pKa = 11.84QDD372 pKa = 3.14TKK374 pKa = 11.64ALGSEE379 pKa = 3.77AGVRR383 pKa = 11.84EE384 pKa = 4.05EE385 pKa = 4.98SSGKK389 pKa = 9.95KK390 pKa = 9.69
MM1 pKa = 7.95RR2 pKa = 11.84LGKK5 pKa = 9.99ILGEE9 pKa = 3.81NRR11 pKa = 11.84SFRR14 pKa = 11.84FYY16 pKa = 9.67WASISLSHH24 pKa = 7.27ISGIMFNVISAYY36 pKa = 9.58ILLLVSPSFYY46 pKa = 10.73SLVVGVTMLVGALIRR61 pKa = 11.84LPSGYY66 pKa = 10.52LSDD69 pKa = 3.74SLDD72 pKa = 3.34RR73 pKa = 11.84RR74 pKa = 11.84KK75 pKa = 10.65ALMLTRR81 pKa = 11.84LSQAVLILLPILSLKK96 pKa = 8.45LTPFSYY102 pKa = 10.96VGFNLLTSLNGPLSAGLVQSVMEE125 pKa = 4.13KK126 pKa = 10.11RR127 pKa = 11.84QVLGGTSLNSLTISISSVVGGLVSLSYY154 pKa = 10.34PVLGITAFLSIASVMRR170 pKa = 11.84LFTVFFIGNVRR181 pKa = 11.84VKK183 pKa = 10.63ARR185 pKa = 11.84KK186 pKa = 9.51DD187 pKa = 3.48EE188 pKa = 4.46PKK190 pKa = 10.8GRR192 pKa = 11.84IPVSLIIYY200 pKa = 7.09PILVFSFLDD209 pKa = 3.95VIPVMMNSLVFVVSSVEE226 pKa = 3.64PSLALVSLLTTLGNGMGAILTGEE249 pKa = 4.73RR250 pKa = 11.84IRR252 pKa = 11.84EE253 pKa = 3.78NSIGIWARR261 pKa = 11.84TGSLLLGLVLVAISLSPLYY280 pKa = 10.24LVYY283 pKa = 10.89VLLVLRR289 pKa = 11.84GILSSIQSISIRR301 pKa = 11.84SDD303 pKa = 2.67LRR305 pKa = 11.84LRR307 pKa = 11.84VSNNLMGRR315 pKa = 11.84TWGLITTISSFVASVVALSYY335 pKa = 11.22SFIVSVTGTRR345 pKa = 11.84FPVLVLGLSLLTLGILMVYY364 pKa = 10.01RR365 pKa = 11.84GPSNRR370 pKa = 11.84QDD372 pKa = 3.14TKK374 pKa = 11.64ALGSEE379 pKa = 3.77AGVRR383 pKa = 11.84EE384 pKa = 4.05EE385 pKa = 4.98SSGKK389 pKa = 9.95KK390 pKa = 9.69
Molecular weight: 42.05 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
640304 |
46 |
1455 |
283.8 |
31.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.192 ± 0.052 | 0.62 ± 0.018 |
4.704 ± 0.046 | 6.931 ± 0.063 |
4.169 ± 0.038 | 7.477 ± 0.042 |
1.427 ± 0.019 | 7.216 ± 0.048 |
6.072 ± 0.054 | 10.573 ± 0.066 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.663 ± 0.023 | 3.812 ± 0.04 |
4.359 ± 0.033 | 2.372 ± 0.029 |
5.612 ± 0.054 | 7.099 ± 0.051 |
5.031 ± 0.051 | 8.684 ± 0.046 |
1.045 ± 0.017 | 3.942 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
