
Novosphingobium sp. PP1Y
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Sphingomonadales; Sphingomonadaceae; Novosphingobium; unclassified Novosphingobium
Average proteome isoelectric point is 6.26
Get precalculated fractions of proteins
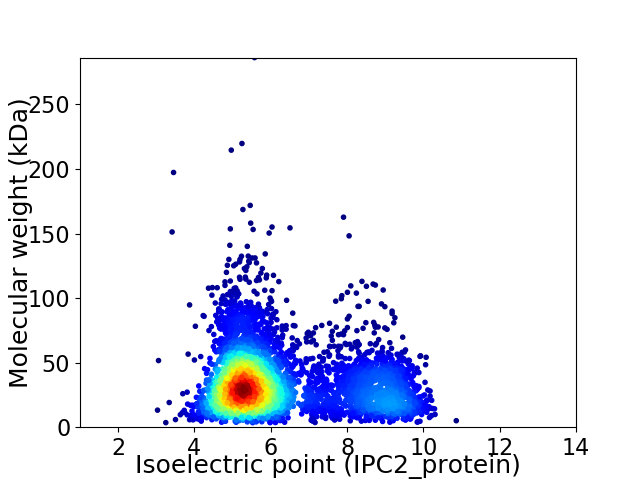
Virtual 2D-PAGE plot for 4624 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|F6IMM9|F6IMM9_9SPHN MFS transporter DHA2 family multidrug resistance protein B OS=Novosphingobium sp. PP1Y OX=702113 GN=PP1Y_AT17977 PE=4 SV=1
MM1 pKa = 7.87ASPALAQDD9 pKa = 3.39APGPITVSGEE19 pKa = 3.86VALVSDD25 pKa = 3.59YY26 pKa = 11.1RR27 pKa = 11.84FRR29 pKa = 11.84GVSQTDD35 pKa = 2.87EE36 pKa = 3.97EE37 pKa = 4.7MAVQGGINVEE47 pKa = 4.33HH48 pKa = 7.2EE49 pKa = 4.25SGLYY53 pKa = 10.22AGTWASNLSGWGTFGGSNMEE73 pKa = 4.1LDD75 pKa = 4.78LYY77 pKa = 11.09AGYY80 pKa = 8.5ATEE83 pKa = 4.45IASGLNVDD91 pKa = 5.44VGMTWFMYY99 pKa = 10.31PGGASDD105 pKa = 3.44TDD107 pKa = 3.75FAEE110 pKa = 4.89VYY112 pKa = 10.5VKK114 pKa = 10.89GSTTFGPADD123 pKa = 3.68FLVGVAYY130 pKa = 10.11APKK133 pKa = 9.49QQALGKK139 pKa = 9.63VYY141 pKa = 11.1NNAASYY147 pKa = 10.61NAGIPDD153 pKa = 3.65NPGAKK158 pKa = 9.35SDD160 pKa = 3.92NVYY163 pKa = 9.66IWSDD167 pKa = 3.15ASVGVPNTPLTLSAHH182 pKa = 6.64LGYY185 pKa = 11.21SNGNSGLGPNGTSIAPTGEE204 pKa = 3.85FLDD207 pKa = 3.82WSLGADD213 pKa = 3.75LALGPVTLGVAYY225 pKa = 10.55VDD227 pKa = 3.9TDD229 pKa = 3.25ISSSDD234 pKa = 3.3AAYY237 pKa = 9.55LQPNFASSKK246 pKa = 9.72DD247 pKa = 3.47GSKK250 pKa = 10.23IADD253 pKa = 3.48STILFSISAGFF264 pKa = 3.32
MM1 pKa = 7.87ASPALAQDD9 pKa = 3.39APGPITVSGEE19 pKa = 3.86VALVSDD25 pKa = 3.59YY26 pKa = 11.1RR27 pKa = 11.84FRR29 pKa = 11.84GVSQTDD35 pKa = 2.87EE36 pKa = 3.97EE37 pKa = 4.7MAVQGGINVEE47 pKa = 4.33HH48 pKa = 7.2EE49 pKa = 4.25SGLYY53 pKa = 10.22AGTWASNLSGWGTFGGSNMEE73 pKa = 4.1LDD75 pKa = 4.78LYY77 pKa = 11.09AGYY80 pKa = 8.5ATEE83 pKa = 4.45IASGLNVDD91 pKa = 5.44VGMTWFMYY99 pKa = 10.31PGGASDD105 pKa = 3.44TDD107 pKa = 3.75FAEE110 pKa = 4.89VYY112 pKa = 10.5VKK114 pKa = 10.89GSTTFGPADD123 pKa = 3.68FLVGVAYY130 pKa = 10.11APKK133 pKa = 9.49QQALGKK139 pKa = 9.63VYY141 pKa = 11.1NNAASYY147 pKa = 10.61NAGIPDD153 pKa = 3.65NPGAKK158 pKa = 9.35SDD160 pKa = 3.92NVYY163 pKa = 9.66IWSDD167 pKa = 3.15ASVGVPNTPLTLSAHH182 pKa = 6.64LGYY185 pKa = 11.21SNGNSGLGPNGTSIAPTGEE204 pKa = 3.85FLDD207 pKa = 3.82WSLGADD213 pKa = 3.75LALGPVTLGVAYY225 pKa = 10.55VDD227 pKa = 3.9TDD229 pKa = 3.25ISSSDD234 pKa = 3.3AAYY237 pKa = 9.55LQPNFASSKK246 pKa = 9.72DD247 pKa = 3.47GSKK250 pKa = 10.23IADD253 pKa = 3.48STILFSISAGFF264 pKa = 3.32
Molecular weight: 27.15 kDa
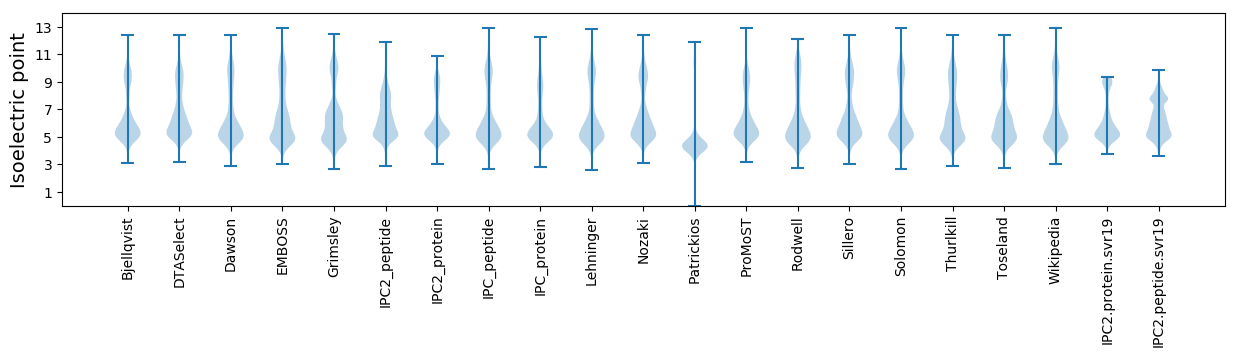
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|F6IPJ2|F6IPJ2_9SPHN dITP/XTP pyrophosphatase OS=Novosphingobium sp. PP1Y OX=702113 GN=PP1Y_AT25532 PE=3 SV=1
MM1 pKa = 7.24EE2 pKa = 5.12TGFCPSRR9 pKa = 11.84LRR11 pKa = 11.84KK12 pKa = 7.94PQKK15 pKa = 10.62SEE17 pKa = 3.52ILPFAGTGVSLIPPPGNLSISSFEE41 pKa = 4.48PGWAPRR47 pKa = 11.84CCSRR51 pKa = 11.84SFRR54 pKa = 11.84SVIWPLAVTVRR65 pKa = 11.84LVIGSFLLFRR75 pKa = 11.84HH76 pKa = 5.55NVRR79 pKa = 11.84IYY81 pKa = 10.69NLTVKK86 pKa = 10.52ASSIAAASLCFTAATIAIARR106 pKa = 11.84PVTPDD111 pKa = 3.13DD112 pKa = 3.8AMRR115 pKa = 11.84AAIHH119 pKa = 6.12PVADD123 pKa = 3.97STDD126 pKa = 3.42MQAWLARR133 pKa = 11.84VPVTPDD139 pKa = 3.74FPPDD143 pKa = 3.32FAVTLRR149 pKa = 11.84GEE151 pKa = 4.07APAFVIEE158 pKa = 4.34MSTTPGCLPCADD170 pKa = 4.95LWTKK174 pKa = 10.61LAVLRR179 pKa = 11.84SRR181 pKa = 11.84YY182 pKa = 8.49GWQIRR187 pKa = 11.84TISGEE192 pKa = 3.94EE193 pKa = 3.53AMLRR197 pKa = 11.84SGRR200 pKa = 11.84LGLPWVGHH208 pKa = 4.87PVAWVRR214 pKa = 11.84PVNDD218 pKa = 4.08PNRR221 pKa = 11.84TVPIAIGTDD230 pKa = 3.19HH231 pKa = 7.13AVNLARR237 pKa = 11.84NAYY240 pKa = 9.28LAAKK244 pKa = 8.5MLTGVRR250 pKa = 11.84VDD252 pKa = 3.3VGVRR256 pKa = 11.84ALSKK260 pKa = 9.26FTGIVGLARR269 pKa = 11.84RR270 pKa = 11.84PASIRR275 pKa = 3.44
MM1 pKa = 7.24EE2 pKa = 5.12TGFCPSRR9 pKa = 11.84LRR11 pKa = 11.84KK12 pKa = 7.94PQKK15 pKa = 10.62SEE17 pKa = 3.52ILPFAGTGVSLIPPPGNLSISSFEE41 pKa = 4.48PGWAPRR47 pKa = 11.84CCSRR51 pKa = 11.84SFRR54 pKa = 11.84SVIWPLAVTVRR65 pKa = 11.84LVIGSFLLFRR75 pKa = 11.84HH76 pKa = 5.55NVRR79 pKa = 11.84IYY81 pKa = 10.69NLTVKK86 pKa = 10.52ASSIAAASLCFTAATIAIARR106 pKa = 11.84PVTPDD111 pKa = 3.13DD112 pKa = 3.8AMRR115 pKa = 11.84AAIHH119 pKa = 6.12PVADD123 pKa = 3.97STDD126 pKa = 3.42MQAWLARR133 pKa = 11.84VPVTPDD139 pKa = 3.74FPPDD143 pKa = 3.32FAVTLRR149 pKa = 11.84GEE151 pKa = 4.07APAFVIEE158 pKa = 4.34MSTTPGCLPCADD170 pKa = 4.95LWTKK174 pKa = 10.61LAVLRR179 pKa = 11.84SRR181 pKa = 11.84YY182 pKa = 8.49GWQIRR187 pKa = 11.84TISGEE192 pKa = 3.94EE193 pKa = 3.53AMLRR197 pKa = 11.84SGRR200 pKa = 11.84LGLPWVGHH208 pKa = 4.87PVAWVRR214 pKa = 11.84PVNDD218 pKa = 4.08PNRR221 pKa = 11.84TVPIAIGTDD230 pKa = 3.19HH231 pKa = 7.13AVNLARR237 pKa = 11.84NAYY240 pKa = 9.28LAAKK244 pKa = 8.5MLTGVRR250 pKa = 11.84VDD252 pKa = 3.3VGVRR256 pKa = 11.84ALSKK260 pKa = 9.26FTGIVGLARR269 pKa = 11.84RR270 pKa = 11.84PASIRR275 pKa = 3.44
Molecular weight: 29.78 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1530888 |
33 |
2582 |
331.1 |
35.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.452 ± 0.047 | 0.913 ± 0.011 |
5.91 ± 0.026 | 5.836 ± 0.034 |
3.679 ± 0.021 | 8.798 ± 0.033 |
2.082 ± 0.017 | 5.017 ± 0.023 |
3.119 ± 0.027 | 9.829 ± 0.043 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.466 ± 0.018 | 2.656 ± 0.022 |
5.2 ± 0.025 | 3.223 ± 0.02 |
7.148 ± 0.033 | 5.663 ± 0.028 |
5.142 ± 0.025 | 7.116 ± 0.029 |
1.443 ± 0.014 | 2.308 ± 0.018 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
