
SAR202 cluster bacterium AC-647-N09_OGT_505m
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Chloroflexi; Chloroflexi incertae sedis; SAR202 cluster
Average proteome isoelectric point is 6.0
Get precalculated fractions of proteins
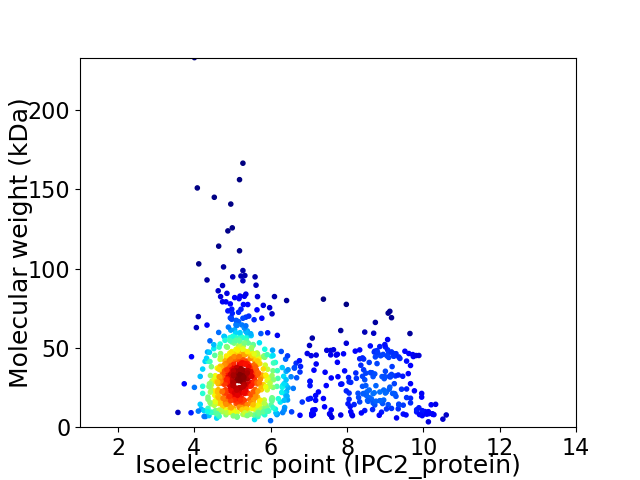
Virtual 2D-PAGE plot for 888 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5N8YAU8|A0A5N8YAU8_9CHLR Homocysteine S-methyltransferase family protein OS=SAR202 cluster bacterium AC-647-N09_OGT_505m OX=2587833 GN=FIM08_00010 PE=4 SV=1
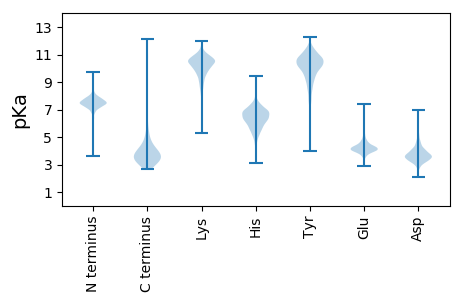
GG1 pKa = 7.04TGTSLIFKK9 pKa = 9.05YY10 pKa = 10.17QGNPTIRR17 pKa = 11.84NNNFVHH23 pKa = 7.15KK24 pKa = 10.5SEE26 pKa = 4.49TYY28 pKa = 10.31LVYY31 pKa = 10.56DD32 pKa = 4.15DD33 pKa = 5.71RR34 pKa = 11.84NVSEE38 pKa = 4.39NATSDD43 pKa = 4.76FEE45 pKa = 4.87NNWWGTTNTTNLDD58 pKa = 3.31ALIYY62 pKa = 10.22DD63 pKa = 3.96WNDD66 pKa = 2.77NATKK70 pKa = 10.66EE71 pKa = 4.35MIDD74 pKa = 3.43YY75 pKa = 9.35TPFLTAPDD83 pKa = 3.8TTAPPSPPANVATQTGPTTISLAWDD108 pKa = 4.03ANPEE112 pKa = 3.98SDD114 pKa = 2.75ITGYY118 pKa = 10.03KK119 pKa = 8.72VHH121 pKa = 7.08YY122 pKa = 8.35DD123 pKa = 3.27TDD125 pKa = 3.86AAGYY129 pKa = 8.68PYY131 pKa = 10.92ANSIDD136 pKa = 3.59IGNVTSYY143 pKa = 9.93TLSGLSTDD151 pKa = 3.39TTYY154 pKa = 9.94YY155 pKa = 9.4TAVSAYY161 pKa = 10.16DD162 pKa = 3.52ADD164 pKa = 4.9GNEE167 pKa = 4.3SWISSNTTATTEE179 pKa = 4.22SVPTALAFSTQPSGATAGNTFTTQPVVVIQGSAEE213 pKa = 4.13NTVTTATDD221 pKa = 3.26SVTISITSGTGGSGATLLGTATVSAVNGVATFTDD255 pKa = 3.6LRR257 pKa = 11.84IDD259 pKa = 3.55KK260 pKa = 10.46AATGYY265 pKa = 8.62TLTATSSSLTLATSASFDD283 pKa = 3.59VSSSATASRR292 pKa = 11.84LVVISEE298 pKa = 4.44PSTTAAGEE306 pKa = 4.36TFVTQPVIQIQDD318 pKa = 3.34VYY320 pKa = 11.16GNIVTSSSASVTVSITSGTGTSGAALSGSAAISAASGVATFSGLGIDD367 pKa = 4.08SAGSNYY373 pKa = 9.87TLTATASGLTSDD385 pKa = 4.19ASSPFDD391 pKa = 3.35VVVAATPIPAMSTWGLIVMALLVSAWMAHH420 pKa = 4.79MARR423 pKa = 11.84RR424 pKa = 11.84GRR426 pKa = 11.84SWIDD430 pKa = 3.01MKK432 pKa = 11.38
GG1 pKa = 7.04TGTSLIFKK9 pKa = 9.05YY10 pKa = 10.17QGNPTIRR17 pKa = 11.84NNNFVHH23 pKa = 7.15KK24 pKa = 10.5SEE26 pKa = 4.49TYY28 pKa = 10.31LVYY31 pKa = 10.56DD32 pKa = 4.15DD33 pKa = 5.71RR34 pKa = 11.84NVSEE38 pKa = 4.39NATSDD43 pKa = 4.76FEE45 pKa = 4.87NNWWGTTNTTNLDD58 pKa = 3.31ALIYY62 pKa = 10.22DD63 pKa = 3.96WNDD66 pKa = 2.77NATKK70 pKa = 10.66EE71 pKa = 4.35MIDD74 pKa = 3.43YY75 pKa = 9.35TPFLTAPDD83 pKa = 3.8TTAPPSPPANVATQTGPTTISLAWDD108 pKa = 4.03ANPEE112 pKa = 3.98SDD114 pKa = 2.75ITGYY118 pKa = 10.03KK119 pKa = 8.72VHH121 pKa = 7.08YY122 pKa = 8.35DD123 pKa = 3.27TDD125 pKa = 3.86AAGYY129 pKa = 8.68PYY131 pKa = 10.92ANSIDD136 pKa = 3.59IGNVTSYY143 pKa = 9.93TLSGLSTDD151 pKa = 3.39TTYY154 pKa = 9.94YY155 pKa = 9.4TAVSAYY161 pKa = 10.16DD162 pKa = 3.52ADD164 pKa = 4.9GNEE167 pKa = 4.3SWISSNTTATTEE179 pKa = 4.22SVPTALAFSTQPSGATAGNTFTTQPVVVIQGSAEE213 pKa = 4.13NTVTTATDD221 pKa = 3.26SVTISITSGTGGSGATLLGTATVSAVNGVATFTDD255 pKa = 3.6LRR257 pKa = 11.84IDD259 pKa = 3.55KK260 pKa = 10.46AATGYY265 pKa = 8.62TLTATSSSLTLATSASFDD283 pKa = 3.59VSSSATASRR292 pKa = 11.84LVVISEE298 pKa = 4.44PSTTAAGEE306 pKa = 4.36TFVTQPVIQIQDD318 pKa = 3.34VYY320 pKa = 11.16GNIVTSSSASVTVSITSGTGTSGAALSGSAAISAASGVATFSGLGIDD367 pKa = 4.08SAGSNYY373 pKa = 9.87TLTATASGLTSDD385 pKa = 4.19ASSPFDD391 pKa = 3.35VVVAATPIPAMSTWGLIVMALLVSAWMAHH420 pKa = 4.79MARR423 pKa = 11.84RR424 pKa = 11.84GRR426 pKa = 11.84SWIDD430 pKa = 3.01MKK432 pKa = 11.38
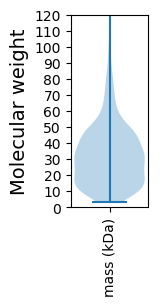
Molecular weight: 44.42 kDa
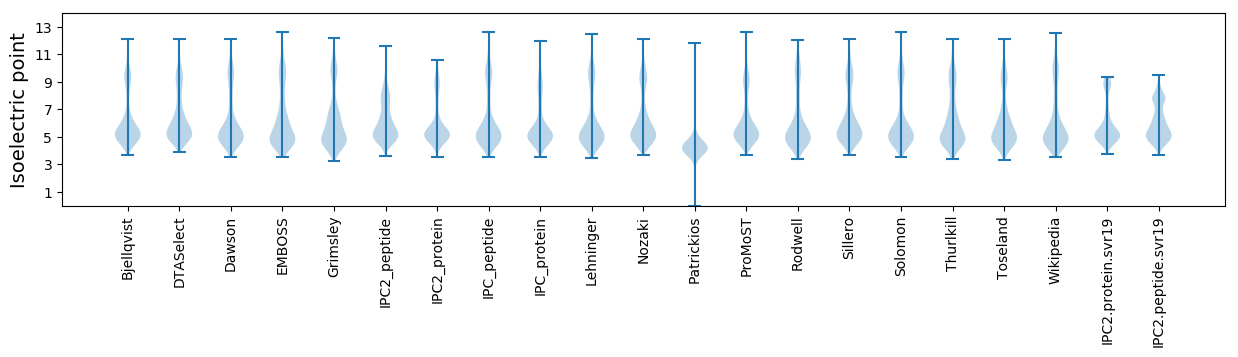
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5N8YDL9|A0A5N8YDL9_9CHLR Molybdenum cofactor biosynthesis protein MoaE OS=SAR202 cluster bacterium AC-647-N09_OGT_505m OX=2587833 GN=FIM08_04185 PE=4 SV=1
MM1 pKa = 7.47PRR3 pKa = 11.84IEE5 pKa = 4.07EE6 pKa = 4.72AIVIPLGPLEE16 pKa = 4.21TWEE19 pKa = 4.34LMRR22 pKa = 11.84DD23 pKa = 3.38LEE25 pKa = 5.48LRR27 pKa = 11.84PRR29 pKa = 11.84WDD31 pKa = 2.98ATIIRR36 pKa = 11.84VGRR39 pKa = 11.84GVVSSDD45 pKa = 3.11AEE47 pKa = 4.22IPRR50 pKa = 11.84LYY52 pKa = 10.95YY53 pKa = 10.37LAPLFLGLRR62 pKa = 11.84WRR64 pKa = 11.84WEE66 pKa = 3.81GEE68 pKa = 4.25YY69 pKa = 10.32ISFQPPNRR77 pKa = 11.84TAVRR81 pKa = 11.84MACGSSLRR89 pKa = 11.84PFKK92 pKa = 11.02SLVGTWVLQSQRR104 pKa = 11.84DD105 pKa = 4.04GTQVRR110 pKa = 11.84MIVSFEE116 pKa = 4.11PRR118 pKa = 11.84WPLPLLGKK126 pKa = 10.33VMGYY130 pKa = 9.78RR131 pKa = 11.84MRR133 pKa = 11.84RR134 pKa = 11.84LLGKK138 pKa = 9.96SLSKK142 pKa = 10.64LRR144 pKa = 11.84AIGTGNHH151 pKa = 6.09SSQDD155 pKa = 3.95SIHH158 pKa = 6.13SS159 pKa = 3.7
MM1 pKa = 7.47PRR3 pKa = 11.84IEE5 pKa = 4.07EE6 pKa = 4.72AIVIPLGPLEE16 pKa = 4.21TWEE19 pKa = 4.34LMRR22 pKa = 11.84DD23 pKa = 3.38LEE25 pKa = 5.48LRR27 pKa = 11.84PRR29 pKa = 11.84WDD31 pKa = 2.98ATIIRR36 pKa = 11.84VGRR39 pKa = 11.84GVVSSDD45 pKa = 3.11AEE47 pKa = 4.22IPRR50 pKa = 11.84LYY52 pKa = 10.95YY53 pKa = 10.37LAPLFLGLRR62 pKa = 11.84WRR64 pKa = 11.84WEE66 pKa = 3.81GEE68 pKa = 4.25YY69 pKa = 10.32ISFQPPNRR77 pKa = 11.84TAVRR81 pKa = 11.84MACGSSLRR89 pKa = 11.84PFKK92 pKa = 11.02SLVGTWVLQSQRR104 pKa = 11.84DD105 pKa = 4.04GTQVRR110 pKa = 11.84MIVSFEE116 pKa = 4.11PRR118 pKa = 11.84WPLPLLGKK126 pKa = 10.33VMGYY130 pKa = 9.78RR131 pKa = 11.84MRR133 pKa = 11.84RR134 pKa = 11.84LLGKK138 pKa = 9.96SLSKK142 pKa = 10.64LRR144 pKa = 11.84AIGTGNHH151 pKa = 6.09SSQDD155 pKa = 3.95SIHH158 pKa = 6.13SS159 pKa = 3.7
Molecular weight: 18.24 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
272155 |
31 |
2239 |
306.5 |
33.75 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.541 ± 0.086 | 0.888 ± 0.026 |
5.336 ± 0.062 | 6.558 ± 0.097 |
3.687 ± 0.056 | 8.471 ± 0.079 |
2.208 ± 0.044 | 6.066 ± 0.075 |
3.39 ± 0.06 | 10.278 ± 0.1 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.751 ± 0.041 | 3.196 ± 0.052 |
4.925 ± 0.052 | 3.585 ± 0.043 |
6.176 ± 0.09 | 6.427 ± 0.065 |
5.628 ± 0.099 | 7.73 ± 0.078 |
1.378 ± 0.037 | 2.783 ± 0.045 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
