
Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) (Halobacterium volcanii)
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Haloferacales; Haloferacaceae; Haloferax; Haloferax volcanii
Average proteome isoelectric point is 5.0
Get precalculated fractions of proteins
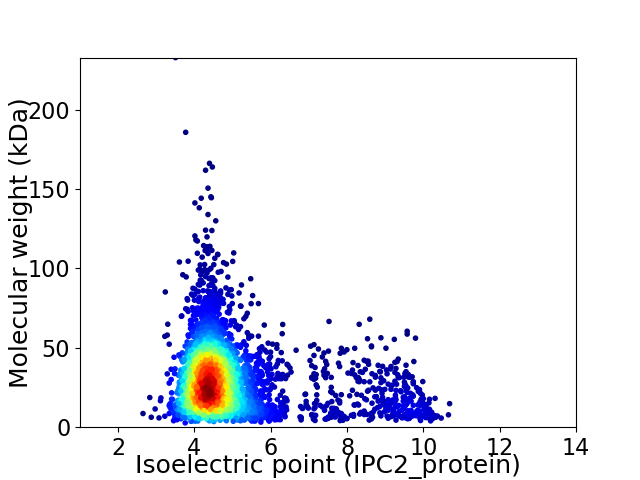
Virtual 2D-PAGE plot for 3911 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D4GTB9|D4GTB9_HALVD Homolog to carboxylate-amine ligase OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=HVO_0744 PE=4 SV=1
MM1 pKa = 7.75SDD3 pKa = 4.14LEE5 pKa = 4.52RR6 pKa = 11.84ICAVGDD12 pKa = 3.78VPDD15 pKa = 4.08DD16 pKa = 3.49TTFLFRR22 pKa = 11.84VRR24 pKa = 11.84AADD27 pKa = 3.82DD28 pKa = 3.68ADD30 pKa = 3.99AEE32 pKa = 4.23EE33 pKa = 5.18LEE35 pKa = 5.04AILVRR40 pKa = 11.84TDD42 pKa = 4.75DD43 pKa = 6.06GIQGWLNYY51 pKa = 9.58CMHH54 pKa = 6.99LTHH57 pKa = 7.02IKK59 pKa = 10.2LDD61 pKa = 3.86KK62 pKa = 11.23GSGAPMRR69 pKa = 11.84NDD71 pKa = 3.07EE72 pKa = 5.3VICQNHH78 pKa = 5.31GAYY81 pKa = 10.36FEE83 pKa = 5.78ADD85 pKa = 2.74TGYY88 pKa = 10.92CNYY91 pKa = 9.67GPCEE95 pKa = 3.94GAVLDD100 pKa = 5.06DD101 pKa = 5.44LDD103 pKa = 3.67ITVDD107 pKa = 3.48GDD109 pKa = 4.25HH110 pKa = 6.74VFLSDD115 pKa = 5.51DD116 pKa = 3.37EE117 pKa = 4.48FDD119 pKa = 3.78YY120 pKa = 11.44VGQGEE125 pKa = 4.79IEE127 pKa = 4.12TDD129 pKa = 3.3PADD132 pKa = 3.97RR133 pKa = 11.84SSSSNVEE140 pKa = 3.96FF141 pKa = 4.89
MM1 pKa = 7.75SDD3 pKa = 4.14LEE5 pKa = 4.52RR6 pKa = 11.84ICAVGDD12 pKa = 3.78VPDD15 pKa = 4.08DD16 pKa = 3.49TTFLFRR22 pKa = 11.84VRR24 pKa = 11.84AADD27 pKa = 3.82DD28 pKa = 3.68ADD30 pKa = 3.99AEE32 pKa = 4.23EE33 pKa = 5.18LEE35 pKa = 5.04AILVRR40 pKa = 11.84TDD42 pKa = 4.75DD43 pKa = 6.06GIQGWLNYY51 pKa = 9.58CMHH54 pKa = 6.99LTHH57 pKa = 7.02IKK59 pKa = 10.2LDD61 pKa = 3.86KK62 pKa = 11.23GSGAPMRR69 pKa = 11.84NDD71 pKa = 3.07EE72 pKa = 5.3VICQNHH78 pKa = 5.31GAYY81 pKa = 10.36FEE83 pKa = 5.78ADD85 pKa = 2.74TGYY88 pKa = 10.92CNYY91 pKa = 9.67GPCEE95 pKa = 3.94GAVLDD100 pKa = 5.06DD101 pKa = 5.44LDD103 pKa = 3.67ITVDD107 pKa = 3.48GDD109 pKa = 4.25HH110 pKa = 6.74VFLSDD115 pKa = 5.51DD116 pKa = 3.37EE117 pKa = 4.48FDD119 pKa = 3.78YY120 pKa = 11.44VGQGEE125 pKa = 4.79IEE127 pKa = 4.12TDD129 pKa = 3.3PADD132 pKa = 3.97RR133 pKa = 11.84SSSSNVEE140 pKa = 3.96FF141 pKa = 4.89
Molecular weight: 15.55 kDa
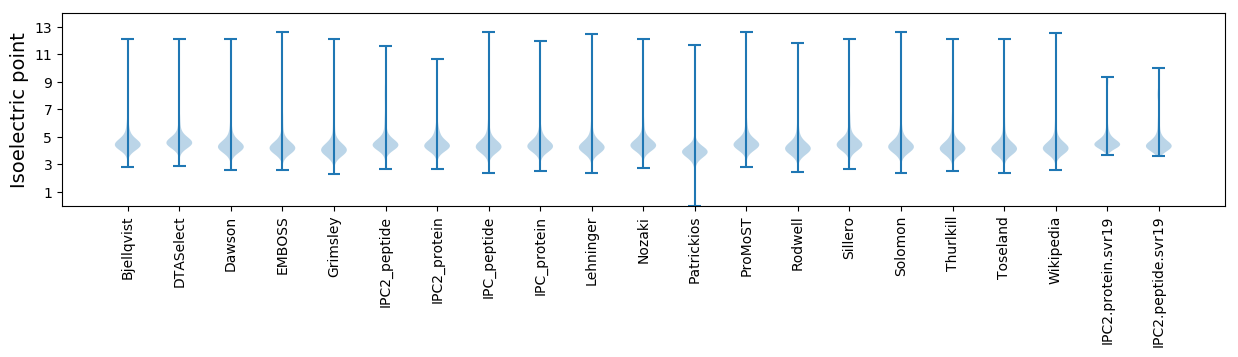
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D4GQM1|D4GQM1_HALVD Polysaccharide deacetylase domain protein OS=Haloferax volcanii (strain ATCC 29605 / DSM 3757 / JCM 8879 / NBRC 14742 / NCIMB 2012 / VKM B-1768 / DS2) OX=309800 GN=HVO_A0192 PE=4 SV=1
MM1 pKa = 7.21IAKK4 pKa = 9.22PKK6 pKa = 6.23TTPRR10 pKa = 11.84LPDD13 pKa = 3.29HH14 pKa = 6.07STPMNRR20 pKa = 11.84ASVARR25 pKa = 11.84VTRR28 pKa = 11.84LVTVVGAAFVLAGLAVQVTTLRR50 pKa = 11.84LGVPHH55 pKa = 6.63LQWGLRR61 pKa = 11.84AASDD65 pKa = 3.98AEE67 pKa = 4.25VLGVGLWWVAVATRR81 pKa = 11.84LVGPRR86 pKa = 11.84RR87 pKa = 11.84TQRR90 pKa = 11.84RR91 pKa = 11.84VGRR94 pKa = 11.84EE95 pKa = 3.5RR96 pKa = 11.84DD97 pKa = 3.47PTRR100 pKa = 11.84QAVYY104 pKa = 8.2VTRR107 pKa = 11.84GLLDD111 pKa = 3.75ALLGRR116 pKa = 11.84AARR119 pKa = 11.84AEE121 pKa = 3.99PSRR124 pKa = 11.84EE125 pKa = 4.04SLSLGTTAAGEE136 pKa = 3.99LDD138 pKa = 3.6GAEE141 pKa = 4.39SLDD144 pKa = 3.18ATRR147 pKa = 11.84PVFSHH152 pKa = 6.86FVHH155 pKa = 6.91PSAGAAVRR163 pKa = 11.84GVFGVDD169 pKa = 3.55LSTAPGRR176 pKa = 11.84TPGQFVSHH184 pKa = 6.99PWGPLAATKK193 pKa = 10.22RR194 pKa = 11.84DD195 pKa = 3.56TLRR198 pKa = 11.84EE199 pKa = 4.09VVLVAVPPWDD209 pKa = 4.52DD210 pKa = 3.5DD211 pKa = 3.79CVAAFDD217 pKa = 3.8RR218 pKa = 11.84AGRR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84PLRR226 pKa = 11.84VVDD229 pKa = 4.89AEE231 pKa = 4.11PCEE234 pKa = 4.49SFDD237 pKa = 4.18EE238 pKa = 5.3AISAAGPAASPPTSSAADD256 pKa = 3.5GG257 pKa = 3.77
MM1 pKa = 7.21IAKK4 pKa = 9.22PKK6 pKa = 6.23TTPRR10 pKa = 11.84LPDD13 pKa = 3.29HH14 pKa = 6.07STPMNRR20 pKa = 11.84ASVARR25 pKa = 11.84VTRR28 pKa = 11.84LVTVVGAAFVLAGLAVQVTTLRR50 pKa = 11.84LGVPHH55 pKa = 6.63LQWGLRR61 pKa = 11.84AASDD65 pKa = 3.98AEE67 pKa = 4.25VLGVGLWWVAVATRR81 pKa = 11.84LVGPRR86 pKa = 11.84RR87 pKa = 11.84TQRR90 pKa = 11.84RR91 pKa = 11.84VGRR94 pKa = 11.84EE95 pKa = 3.5RR96 pKa = 11.84DD97 pKa = 3.47PTRR100 pKa = 11.84QAVYY104 pKa = 8.2VTRR107 pKa = 11.84GLLDD111 pKa = 3.75ALLGRR116 pKa = 11.84AARR119 pKa = 11.84AEE121 pKa = 3.99PSRR124 pKa = 11.84EE125 pKa = 4.04SLSLGTTAAGEE136 pKa = 3.99LDD138 pKa = 3.6GAEE141 pKa = 4.39SLDD144 pKa = 3.18ATRR147 pKa = 11.84PVFSHH152 pKa = 6.86FVHH155 pKa = 6.91PSAGAAVRR163 pKa = 11.84GVFGVDD169 pKa = 3.55LSTAPGRR176 pKa = 11.84TPGQFVSHH184 pKa = 6.99PWGPLAATKK193 pKa = 10.22RR194 pKa = 11.84DD195 pKa = 3.56TLRR198 pKa = 11.84EE199 pKa = 4.09VVLVAVPPWDD209 pKa = 4.52DD210 pKa = 3.5DD211 pKa = 3.79CVAAFDD217 pKa = 3.8RR218 pKa = 11.84AGRR221 pKa = 11.84RR222 pKa = 11.84RR223 pKa = 11.84PLRR226 pKa = 11.84VVDD229 pKa = 4.89AEE231 pKa = 4.11PCEE234 pKa = 4.49SFDD237 pKa = 4.18EE238 pKa = 5.3AISAAGPAASPPTSSAADD256 pKa = 3.5GG257 pKa = 3.77
Molecular weight: 27.12 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1114463 |
27 |
2257 |
285.0 |
30.92 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.09 ± 0.063 | 0.705 ± 0.013 |
8.472 ± 0.055 | 8.0 ± 0.061 |
3.488 ± 0.03 | 8.545 ± 0.045 |
1.966 ± 0.02 | 3.85 ± 0.032 |
2.005 ± 0.029 | 9.05 ± 0.05 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.794 ± 0.017 | 2.382 ± 0.024 |
4.597 ± 0.025 | 2.381 ± 0.023 |
6.61 ± 0.044 | 5.827 ± 0.033 |
6.199 ± 0.036 | 9.236 ± 0.049 |
1.122 ± 0.015 | 2.682 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
