
Atlantic salmon paramyxovirus (isolate -/Norway/Yrkje371/1995) (ASPV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Aquaparamyxovirus; Salmon aquaparamyxovirus
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
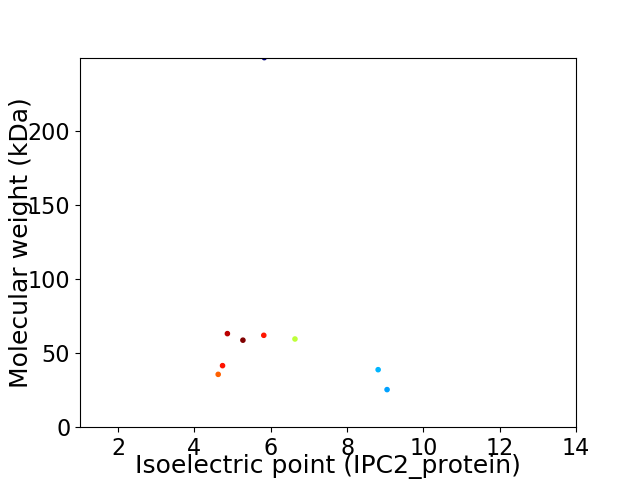
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|B2KSJ3|B2KSJ3_ASPVR Isoform of B2BX74 W protein OS=Atlantic salmon paramyxovirus (isolate -/Norway/Yrkje371/1995) OX=1283346 GN=P/V/C PE=4 SV=1
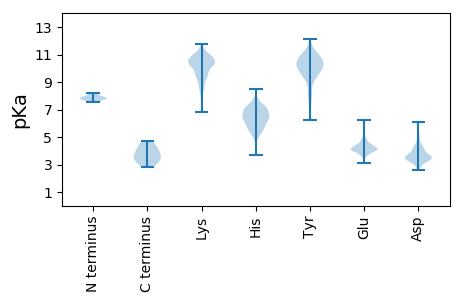
MM1 pKa = 7.87AEE3 pKa = 3.84QKK5 pKa = 10.56GYY7 pKa = 10.2PNIKK11 pKa = 10.65LNDD14 pKa = 3.4EE15 pKa = 4.45FLNEE19 pKa = 3.94GLNILGCFLRR29 pKa = 11.84GNAQTAPTQEE39 pKa = 4.2AQGHH43 pKa = 5.9KK44 pKa = 10.43GGEE47 pKa = 4.13NDD49 pKa = 3.34TGASQKK55 pKa = 10.92APTPTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84NSLSGPSDD71 pKa = 3.47RR72 pKa = 11.84EE73 pKa = 3.72MRR75 pKa = 11.84SQRR78 pKa = 11.84EE79 pKa = 3.68ADD81 pKa = 3.31VDD83 pKa = 3.47KK84 pKa = 10.18WDD86 pKa = 3.45HH87 pKa = 6.23SGVYY91 pKa = 9.27SHH93 pKa = 6.83GLRR96 pKa = 11.84GQLDD100 pKa = 3.38KK101 pKa = 10.93STCEE105 pKa = 3.74EE106 pKa = 4.17FGRR109 pKa = 11.84GQDD112 pKa = 3.63GNTGGLNLCGPDD124 pKa = 3.22VVLNQGPPHH133 pKa = 7.38KK134 pKa = 10.53DD135 pKa = 2.77RR136 pKa = 11.84ALGGIADD143 pKa = 4.33PGKK146 pKa = 9.15KK147 pKa = 7.34THH149 pKa = 7.05LEE151 pKa = 3.95PADD154 pKa = 4.16LEE156 pKa = 4.75DD157 pKa = 4.58GADD160 pKa = 3.67EE161 pKa = 4.29EE162 pKa = 4.69SLPRR166 pKa = 11.84EE167 pKa = 4.09TGGRR171 pKa = 11.84APAGRR176 pKa = 11.84SMEE179 pKa = 4.25GGFTLDD185 pKa = 4.83RR186 pKa = 11.84RR187 pKa = 11.84GSPCKK192 pKa = 9.41PARR195 pKa = 11.84KK196 pKa = 9.41ADD198 pKa = 3.67FRR200 pKa = 11.84EE201 pKa = 4.79GTDD204 pKa = 5.39NPDD207 pKa = 2.72TWSDD211 pKa = 3.23PGTTFMRR218 pKa = 11.84KK219 pKa = 8.53ARR221 pKa = 11.84AMNPQSSASPYY232 pKa = 10.7LEE234 pKa = 4.09EE235 pKa = 3.98QLKK238 pKa = 10.89AEE240 pKa = 4.38KK241 pKa = 10.46AQLLYY246 pKa = 10.46QAEE249 pKa = 4.61DD250 pKa = 3.51NNEE253 pKa = 4.06DD254 pKa = 3.69KK255 pKa = 11.37SEE257 pKa = 4.06GSAPEE262 pKa = 4.11AEE264 pKa = 4.12EE265 pKa = 4.07DD266 pKa = 3.58QEE268 pKa = 4.34EE269 pKa = 4.38EE270 pKa = 4.26EE271 pKa = 4.55EE272 pKa = 3.99EE273 pKa = 4.32DD274 pKa = 5.19EE275 pKa = 4.93IVEE278 pKa = 5.38DD279 pKa = 4.26PDD281 pKa = 3.85SDD283 pKa = 3.72GGEE286 pKa = 4.12TSGLLDD292 pKa = 3.4AHH294 pKa = 6.4NARR297 pKa = 11.84AVDD300 pKa = 3.28AGNRR304 pKa = 11.84IRR306 pKa = 11.84DD307 pKa = 3.35QIYY310 pKa = 9.86DD311 pKa = 3.43VKK313 pKa = 10.41TRR315 pKa = 11.84EE316 pKa = 4.07EE317 pKa = 3.97EE318 pKa = 3.76EE319 pKa = 3.74ARR321 pKa = 11.84QVGKK325 pKa = 10.75KK326 pKa = 9.77GAA328 pKa = 3.54
MM1 pKa = 7.87AEE3 pKa = 3.84QKK5 pKa = 10.56GYY7 pKa = 10.2PNIKK11 pKa = 10.65LNDD14 pKa = 3.4EE15 pKa = 4.45FLNEE19 pKa = 3.94GLNILGCFLRR29 pKa = 11.84GNAQTAPTQEE39 pKa = 4.2AQGHH43 pKa = 5.9KK44 pKa = 10.43GGEE47 pKa = 4.13NDD49 pKa = 3.34TGASQKK55 pKa = 10.92APTPTRR61 pKa = 11.84RR62 pKa = 11.84RR63 pKa = 11.84NSLSGPSDD71 pKa = 3.47RR72 pKa = 11.84EE73 pKa = 3.72MRR75 pKa = 11.84SQRR78 pKa = 11.84EE79 pKa = 3.68ADD81 pKa = 3.31VDD83 pKa = 3.47KK84 pKa = 10.18WDD86 pKa = 3.45HH87 pKa = 6.23SGVYY91 pKa = 9.27SHH93 pKa = 6.83GLRR96 pKa = 11.84GQLDD100 pKa = 3.38KK101 pKa = 10.93STCEE105 pKa = 3.74EE106 pKa = 4.17FGRR109 pKa = 11.84GQDD112 pKa = 3.63GNTGGLNLCGPDD124 pKa = 3.22VVLNQGPPHH133 pKa = 7.38KK134 pKa = 10.53DD135 pKa = 2.77RR136 pKa = 11.84ALGGIADD143 pKa = 4.33PGKK146 pKa = 9.15KK147 pKa = 7.34THH149 pKa = 7.05LEE151 pKa = 3.95PADD154 pKa = 4.16LEE156 pKa = 4.75DD157 pKa = 4.58GADD160 pKa = 3.67EE161 pKa = 4.29EE162 pKa = 4.69SLPRR166 pKa = 11.84EE167 pKa = 4.09TGGRR171 pKa = 11.84APAGRR176 pKa = 11.84SMEE179 pKa = 4.25GGFTLDD185 pKa = 4.83RR186 pKa = 11.84RR187 pKa = 11.84GSPCKK192 pKa = 9.41PARR195 pKa = 11.84KK196 pKa = 9.41ADD198 pKa = 3.67FRR200 pKa = 11.84EE201 pKa = 4.79GTDD204 pKa = 5.39NPDD207 pKa = 2.72TWSDD211 pKa = 3.23PGTTFMRR218 pKa = 11.84KK219 pKa = 8.53ARR221 pKa = 11.84AMNPQSSASPYY232 pKa = 10.7LEE234 pKa = 4.09EE235 pKa = 3.98QLKK238 pKa = 10.89AEE240 pKa = 4.38KK241 pKa = 10.46AQLLYY246 pKa = 10.46QAEE249 pKa = 4.61DD250 pKa = 3.51NNEE253 pKa = 4.06DD254 pKa = 3.69KK255 pKa = 11.37SEE257 pKa = 4.06GSAPEE262 pKa = 4.11AEE264 pKa = 4.12EE265 pKa = 4.07DD266 pKa = 3.58QEE268 pKa = 4.34EE269 pKa = 4.38EE270 pKa = 4.26EE271 pKa = 4.55EE272 pKa = 3.99EE273 pKa = 4.32DD274 pKa = 5.19EE275 pKa = 4.93IVEE278 pKa = 5.38DD279 pKa = 4.26PDD281 pKa = 3.85SDD283 pKa = 3.72GGEE286 pKa = 4.12TSGLLDD292 pKa = 3.4AHH294 pKa = 6.4NARR297 pKa = 11.84AVDD300 pKa = 3.28AGNRR304 pKa = 11.84IRR306 pKa = 11.84DD307 pKa = 3.35QIYY310 pKa = 9.86DD311 pKa = 3.43VKK313 pKa = 10.41TRR315 pKa = 11.84EE316 pKa = 4.07EE317 pKa = 3.97EE318 pKa = 3.76EE319 pKa = 3.74ARR321 pKa = 11.84QVGKK325 pKa = 10.75KK326 pKa = 9.77GAA328 pKa = 3.54
Molecular weight: 35.68 kDa
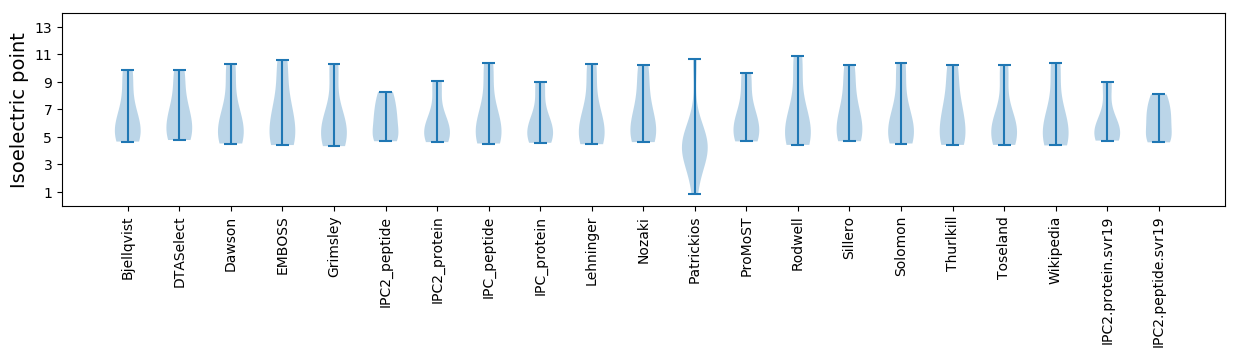
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|B2KSJ2|B2KSJ2_ASPVR Isoform of B2BX74 V protein OS=Atlantic salmon paramyxovirus (isolate -/Norway/Yrkje371/1995) OX=1283346 GN=P/V/C PE=4 SV=1
MM1 pKa = 7.77SSILDD6 pKa = 4.23KK7 pKa = 10.78IKK9 pKa = 10.08RR10 pKa = 11.84WQNKK14 pKa = 8.74RR15 pKa = 11.84ATQTSSSMMSSLTKK29 pKa = 10.32VSTSSGVSSEE39 pKa = 4.38EE40 pKa = 4.09MPKK43 pKa = 10.49LPQPKK48 pKa = 8.89KK49 pKa = 9.86HH50 pKa = 6.62RR51 pKa = 11.84DD52 pKa = 3.15TRR54 pKa = 11.84VVKK57 pKa = 9.07MIPVPLKK64 pKa = 10.56RR65 pKa = 11.84PQLPPGEE72 pKa = 4.28GTVLVVHH79 pKa = 7.05PIGKK83 pKa = 9.46CDD85 pKa = 3.56PRR87 pKa = 11.84EE88 pKa = 4.02KK89 pKa = 10.7RR90 pKa = 11.84MSTSGIIVAYY100 pKa = 9.6IATDD104 pKa = 3.11SGANSIRR111 pKa = 11.84ALVKK115 pKa = 10.35SLEE118 pKa = 4.23EE119 pKa = 4.07VKK121 pKa = 10.11MEE123 pKa = 4.11TPEE126 pKa = 4.47DD127 pKa = 4.08SIFVDD132 pKa = 4.63QMLCSTRR139 pKa = 11.84VLLIRR144 pKa = 11.84IEE146 pKa = 4.07HH147 pKa = 5.43WVEE150 pKa = 3.27LLTRR154 pKa = 11.84GRR156 pKa = 11.84RR157 pKa = 11.84PTWNLLISRR166 pKa = 11.84MALMKK171 pKa = 10.19RR172 pKa = 11.84VSQGKK177 pKa = 8.22PVEE180 pKa = 4.06EE181 pKa = 4.46LLLDD185 pKa = 4.11EE186 pKa = 4.55AWKK189 pKa = 10.85VVLPWIEE196 pKa = 4.18GEE198 pKa = 4.39VRR200 pKa = 11.84ASLPAKK206 pKa = 10.05QISEE210 pKa = 4.31KK211 pKa = 10.88GQTTPTPGQTQEE223 pKa = 3.96RR224 pKa = 11.84LLL226 pKa = 4.04
MM1 pKa = 7.77SSILDD6 pKa = 4.23KK7 pKa = 10.78IKK9 pKa = 10.08RR10 pKa = 11.84WQNKK14 pKa = 8.74RR15 pKa = 11.84ATQTSSSMMSSLTKK29 pKa = 10.32VSTSSGVSSEE39 pKa = 4.38EE40 pKa = 4.09MPKK43 pKa = 10.49LPQPKK48 pKa = 8.89KK49 pKa = 9.86HH50 pKa = 6.62RR51 pKa = 11.84DD52 pKa = 3.15TRR54 pKa = 11.84VVKK57 pKa = 9.07MIPVPLKK64 pKa = 10.56RR65 pKa = 11.84PQLPPGEE72 pKa = 4.28GTVLVVHH79 pKa = 7.05PIGKK83 pKa = 9.46CDD85 pKa = 3.56PRR87 pKa = 11.84EE88 pKa = 4.02KK89 pKa = 10.7RR90 pKa = 11.84MSTSGIIVAYY100 pKa = 9.6IATDD104 pKa = 3.11SGANSIRR111 pKa = 11.84ALVKK115 pKa = 10.35SLEE118 pKa = 4.23EE119 pKa = 4.07VKK121 pKa = 10.11MEE123 pKa = 4.11TPEE126 pKa = 4.47DD127 pKa = 4.08SIFVDD132 pKa = 4.63QMLCSTRR139 pKa = 11.84VLLIRR144 pKa = 11.84IEE146 pKa = 4.07HH147 pKa = 5.43WVEE150 pKa = 3.27LLTRR154 pKa = 11.84GRR156 pKa = 11.84RR157 pKa = 11.84PTWNLLISRR166 pKa = 11.84MALMKK171 pKa = 10.19RR172 pKa = 11.84VSQGKK177 pKa = 8.22PVEE180 pKa = 4.06EE181 pKa = 4.46LLLDD185 pKa = 4.11EE186 pKa = 4.55AWKK189 pKa = 10.85VVLPWIEE196 pKa = 4.18GEE198 pKa = 4.39VRR200 pKa = 11.84ASLPAKK206 pKa = 10.05QISEE210 pKa = 4.31KK211 pKa = 10.88GQTTPTPGQTQEE223 pKa = 3.96RR224 pKa = 11.84LLL226 pKa = 4.04
Molecular weight: 25.38 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
5728 |
226 |
2209 |
636.4 |
70.5 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.529 ± 0.455 | 1.92 ± 0.27 |
5.761 ± 0.497 | 7.071 ± 0.821 |
2.968 ± 0.539 | 7.49 ± 0.697 |
1.641 ± 0.145 | 5.901 ± 0.596 |
5.709 ± 0.41 | 8.834 ± 0.688 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.898 ± 0.399 | 4.452 ± 0.376 |
5.045 ± 0.728 | 3.352 ± 0.259 |
5.499 ± 0.401 | 7.891 ± 0.399 |
6.494 ± 0.423 | 6.39 ± 0.595 |
1.17 ± 0.156 | 2.985 ± 0.313 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
