
Leptolyngbya sp. NIES-2104
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Cyanobacteria/Melainabacteria group; Cyanobacteria; Synechococcales; Leptolyngbyaceae; Leptolyngbya; unclassified Leptolyngbya
Average proteome isoelectric point is 6.33
Get precalculated fractions of proteins
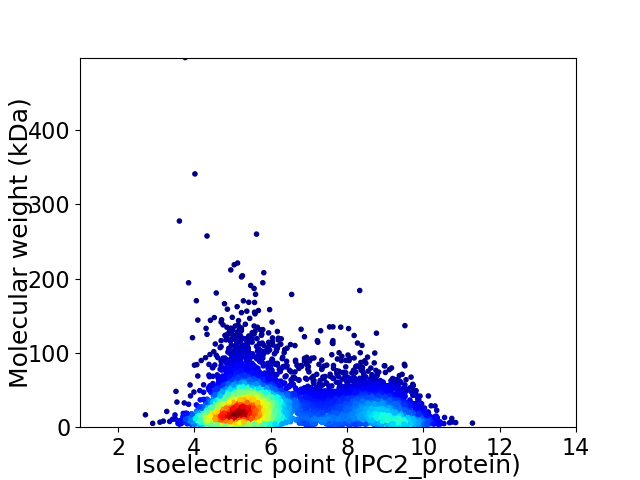
Virtual 2D-PAGE plot for 6684 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0P4V7M8|A0A0P4V7M8_9CYAN Uncharacterized protein OS=Leptolyngbya sp. NIES-2104 OX=1552121 GN=NIES2104_64880 PE=4 SV=1
MM1 pKa = 7.33IAIDD5 pKa = 3.62SFEE8 pKa = 4.91NGGSWTRR15 pKa = 11.84LRR17 pKa = 11.84DD18 pKa = 4.1FGAYY22 pKa = 8.21EE23 pKa = 3.85QANNPDD29 pKa = 3.6GQNVDD34 pKa = 3.34TNLYY38 pKa = 10.69DD39 pKa = 5.56LLINDD44 pKa = 3.27NTAYY48 pKa = 10.78VVDD51 pKa = 4.88AGANDD56 pKa = 4.56LLSQRR61 pKa = 11.84AFSSEE66 pKa = 4.01VNLEE70 pKa = 3.9AVLPTRR76 pKa = 11.84KK77 pKa = 8.52ATNPFTGEE85 pKa = 4.3AIVQQSVPTATAVGPDD101 pKa = 3.08GALYY105 pKa = 10.58VSEE108 pKa = 4.05LTGFPFQPEE117 pKa = 3.86TAQVYY122 pKa = 10.1RR123 pKa = 11.84INTEE127 pKa = 4.05GQPEE131 pKa = 4.68VYY133 pKa = 10.46AGGSTNIVDD142 pKa = 4.74LAFDD146 pKa = 3.6QSGGLYY152 pKa = 9.89VLEE155 pKa = 4.21YY156 pKa = 10.68DD157 pKa = 5.22ANGLLNEE164 pKa = 4.53GDD166 pKa = 3.92AGALIYY172 pKa = 10.21LSPDD176 pKa = 2.58GQMRR180 pKa = 11.84STITDD185 pKa = 3.62DD186 pKa = 3.82LVSPTGLEE194 pKa = 3.6IGADD198 pKa = 3.2GDD200 pKa = 4.53LYY202 pKa = 11.36VSNKK206 pKa = 9.77GFVAGQGEE214 pKa = 4.63VLRR217 pKa = 11.84LSLEE221 pKa = 4.31GKK223 pKa = 9.76SFNPDD228 pKa = 4.33PICDD232 pKa = 3.45QVKK235 pKa = 10.32YY236 pKa = 8.99YY237 pKa = 7.27TTTISADD244 pKa = 3.26GDD246 pKa = 4.0PADD249 pKa = 3.49VYY251 pKa = 11.62YY252 pKa = 10.14PVLPNSNPDD261 pKa = 3.49QLPIALMLQGALVDD275 pKa = 3.62KK276 pKa = 10.98ADD278 pKa = 3.57YY279 pKa = 10.63SSYY282 pKa = 10.81AEE284 pKa = 4.11EE285 pKa = 3.94VASYY289 pKa = 10.43GFVVVVPNNEE299 pKa = 4.01RR300 pKa = 11.84TLTAPPGSPEE310 pKa = 3.83PYY312 pKa = 8.55ITGFLADD319 pKa = 3.79QQQVTDD325 pKa = 3.6VLAQMRR331 pKa = 11.84IEE333 pKa = 4.34DD334 pKa = 4.29TDD336 pKa = 3.92TNSPISGIVDD346 pKa = 3.64TSKK349 pKa = 11.15LGLLGHH355 pKa = 6.19SFGGYY360 pKa = 10.13AGLASIQDD368 pKa = 3.51INDD371 pKa = 3.87PAVSSGNYY379 pKa = 7.01TRR381 pKa = 11.84PSEE384 pKa = 4.02LKK386 pKa = 10.35AGIFYY391 pKa = 7.65GTNFQTPPSSRR402 pKa = 11.84VFPPIDD408 pKa = 3.6NQDD411 pKa = 3.28IPVGLIAGTLDD422 pKa = 3.24GVADD426 pKa = 4.33LGEE429 pKa = 4.16VASTYY434 pKa = 11.17VKK436 pKa = 10.89VQDD439 pKa = 4.07PSKK442 pKa = 11.3ALITVEE448 pKa = 4.13GANHH452 pKa = 6.15YY453 pKa = 10.91GITNQDD459 pKa = 2.96SDD461 pKa = 4.33RR462 pKa = 11.84DD463 pKa = 4.41SVCPTLDD470 pKa = 3.02QATANGATLAVSASQRR486 pKa = 11.84GRR488 pKa = 11.84WSGLFLRR495 pKa = 11.84EE496 pKa = 3.92LRR498 pKa = 11.84CTGTCSHH505 pKa = 7.19LLGDD509 pKa = 4.96PGAFDD514 pKa = 3.85YY515 pKa = 11.19VYY517 pKa = 9.77KK518 pKa = 10.61TGGDD522 pKa = 3.75LNPTVNVTSQTSGII536 pKa = 3.94
MM1 pKa = 7.33IAIDD5 pKa = 3.62SFEE8 pKa = 4.91NGGSWTRR15 pKa = 11.84LRR17 pKa = 11.84DD18 pKa = 4.1FGAYY22 pKa = 8.21EE23 pKa = 3.85QANNPDD29 pKa = 3.6GQNVDD34 pKa = 3.34TNLYY38 pKa = 10.69DD39 pKa = 5.56LLINDD44 pKa = 3.27NTAYY48 pKa = 10.78VVDD51 pKa = 4.88AGANDD56 pKa = 4.56LLSQRR61 pKa = 11.84AFSSEE66 pKa = 4.01VNLEE70 pKa = 3.9AVLPTRR76 pKa = 11.84KK77 pKa = 8.52ATNPFTGEE85 pKa = 4.3AIVQQSVPTATAVGPDD101 pKa = 3.08GALYY105 pKa = 10.58VSEE108 pKa = 4.05LTGFPFQPEE117 pKa = 3.86TAQVYY122 pKa = 10.1RR123 pKa = 11.84INTEE127 pKa = 4.05GQPEE131 pKa = 4.68VYY133 pKa = 10.46AGGSTNIVDD142 pKa = 4.74LAFDD146 pKa = 3.6QSGGLYY152 pKa = 9.89VLEE155 pKa = 4.21YY156 pKa = 10.68DD157 pKa = 5.22ANGLLNEE164 pKa = 4.53GDD166 pKa = 3.92AGALIYY172 pKa = 10.21LSPDD176 pKa = 2.58GQMRR180 pKa = 11.84STITDD185 pKa = 3.62DD186 pKa = 3.82LVSPTGLEE194 pKa = 3.6IGADD198 pKa = 3.2GDD200 pKa = 4.53LYY202 pKa = 11.36VSNKK206 pKa = 9.77GFVAGQGEE214 pKa = 4.63VLRR217 pKa = 11.84LSLEE221 pKa = 4.31GKK223 pKa = 9.76SFNPDD228 pKa = 4.33PICDD232 pKa = 3.45QVKK235 pKa = 10.32YY236 pKa = 8.99YY237 pKa = 7.27TTTISADD244 pKa = 3.26GDD246 pKa = 4.0PADD249 pKa = 3.49VYY251 pKa = 11.62YY252 pKa = 10.14PVLPNSNPDD261 pKa = 3.49QLPIALMLQGALVDD275 pKa = 3.62KK276 pKa = 10.98ADD278 pKa = 3.57YY279 pKa = 10.63SSYY282 pKa = 10.81AEE284 pKa = 4.11EE285 pKa = 3.94VASYY289 pKa = 10.43GFVVVVPNNEE299 pKa = 4.01RR300 pKa = 11.84TLTAPPGSPEE310 pKa = 3.83PYY312 pKa = 8.55ITGFLADD319 pKa = 3.79QQQVTDD325 pKa = 3.6VLAQMRR331 pKa = 11.84IEE333 pKa = 4.34DD334 pKa = 4.29TDD336 pKa = 3.92TNSPISGIVDD346 pKa = 3.64TSKK349 pKa = 11.15LGLLGHH355 pKa = 6.19SFGGYY360 pKa = 10.13AGLASIQDD368 pKa = 3.51INDD371 pKa = 3.87PAVSSGNYY379 pKa = 7.01TRR381 pKa = 11.84PSEE384 pKa = 4.02LKK386 pKa = 10.35AGIFYY391 pKa = 7.65GTNFQTPPSSRR402 pKa = 11.84VFPPIDD408 pKa = 3.6NQDD411 pKa = 3.28IPVGLIAGTLDD422 pKa = 3.24GVADD426 pKa = 4.33LGEE429 pKa = 4.16VASTYY434 pKa = 11.17VKK436 pKa = 10.89VQDD439 pKa = 4.07PSKK442 pKa = 11.3ALITVEE448 pKa = 4.13GANHH452 pKa = 6.15YY453 pKa = 10.91GITNQDD459 pKa = 2.96SDD461 pKa = 4.33RR462 pKa = 11.84DD463 pKa = 4.41SVCPTLDD470 pKa = 3.02QATANGATLAVSASQRR486 pKa = 11.84GRR488 pKa = 11.84WSGLFLRR495 pKa = 11.84EE496 pKa = 3.92LRR498 pKa = 11.84CTGTCSHH505 pKa = 7.19LLGDD509 pKa = 4.96PGAFDD514 pKa = 3.85YY515 pKa = 11.19VYY517 pKa = 9.77KK518 pKa = 10.61TGGDD522 pKa = 3.75LNPTVNVTSQTSGII536 pKa = 3.94
Molecular weight: 56.8 kDa
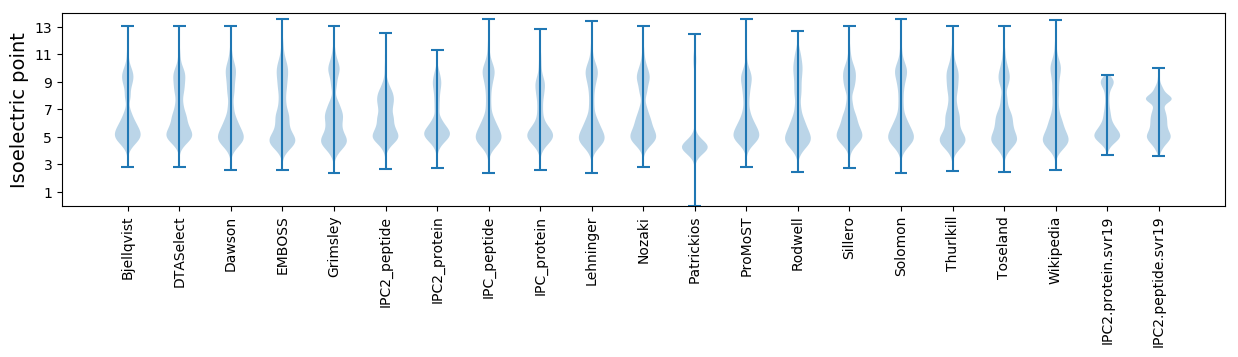
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0P4UTS3|A0A0P4UTS3_9CYAN Uncharacterized protein OS=Leptolyngbya sp. NIES-2104 OX=1552121 GN=NIES2104_23520 PE=4 SV=1
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.4NGQKK30 pKa = 10.06VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2KK38 pKa = 10.15GRR40 pKa = 11.84LRR42 pKa = 11.84LSVSSSS48 pKa = 2.84
MM1 pKa = 7.55TKK3 pKa = 9.01RR4 pKa = 11.84TLGGTSRR11 pKa = 11.84KK12 pKa = 8.87RR13 pKa = 11.84RR14 pKa = 11.84RR15 pKa = 11.84VSGFRR20 pKa = 11.84ARR22 pKa = 11.84MRR24 pKa = 11.84TKK26 pKa = 10.4NGQKK30 pKa = 10.06VIQARR35 pKa = 11.84RR36 pKa = 11.84KK37 pKa = 9.2KK38 pKa = 10.15GRR40 pKa = 11.84LRR42 pKa = 11.84LSVSSSS48 pKa = 2.84
Molecular weight: 5.55 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1864961 |
37 |
4762 |
279.0 |
31.06 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.75 ± 0.03 | 0.935 ± 0.012 |
5.182 ± 0.031 | 6.202 ± 0.036 |
4.023 ± 0.024 | 6.635 ± 0.037 |
1.827 ± 0.018 | 6.436 ± 0.023 |
3.923 ± 0.026 | 10.781 ± 0.034 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.959 ± 0.015 | 3.735 ± 0.025 |
4.797 ± 0.023 | 5.478 ± 0.029 |
6.043 ± 0.024 | 6.653 ± 0.028 |
5.731 ± 0.035 | 6.728 ± 0.028 |
1.442 ± 0.013 | 2.741 ± 0.02 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
