
Thalassoglobus neptunius
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Planctomycetales; Planctomycetaceae; Thalassoglobus
Average proteome isoelectric point is 5.88
Get precalculated fractions of proteins
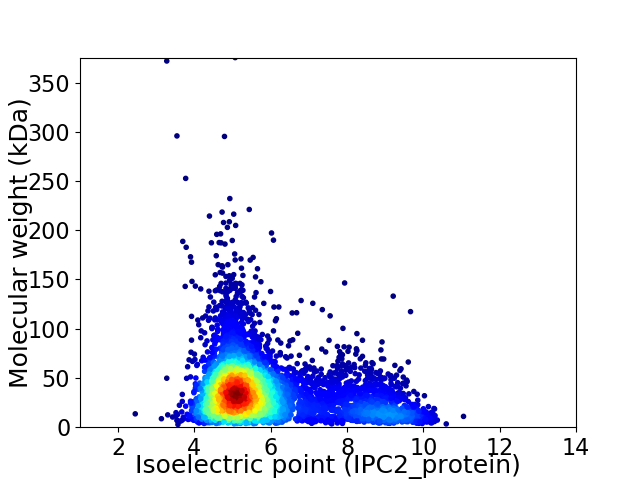
Virtual 2D-PAGE plot for 5460 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5C5X3V5|A0A5C5X3V5_9PLAN Uncharacterized protein OS=Thalassoglobus neptunius OX=1938619 GN=KOR42_03580 PE=4 SV=1
MM1 pKa = 7.31SRR3 pKa = 11.84FAFVFAVLCLTGSFVQAQTTLYY25 pKa = 10.61SDD27 pKa = 3.98SFTNDD32 pKa = 2.28GSAVNLPSWDD42 pKa = 3.52TVEE45 pKa = 5.49NGGVWWNDD53 pKa = 3.1GSDD56 pKa = 3.54STPWVTWSGTDD67 pKa = 3.48SVAHH71 pKa = 6.51HH72 pKa = 6.67SLDD75 pKa = 4.27AIVPGSEE82 pKa = 3.79HH83 pKa = 7.35SGIDD87 pKa = 3.37FGAADD92 pKa = 3.67VDD94 pKa = 3.58ITFEE98 pKa = 4.21FNYY101 pKa = 9.12GTGTTASYY109 pKa = 11.51DD110 pKa = 3.69SIFWIRR116 pKa = 11.84ANSDD120 pKa = 2.77RR121 pKa = 11.84SEE123 pKa = 3.99ALGIYY128 pKa = 10.03HH129 pKa = 6.54SVVDD133 pKa = 5.33DD134 pKa = 4.03EE135 pKa = 4.68FTLVDD140 pKa = 4.12EE141 pKa = 5.38DD142 pKa = 4.36EE143 pKa = 4.56VDD145 pKa = 3.8LGEE148 pKa = 4.34TPVASGLTTANTYY161 pKa = 7.02YY162 pKa = 10.38TIRR165 pKa = 11.84ILLSGSKK172 pKa = 9.94ISLWVDD178 pKa = 3.18DD179 pKa = 4.15VLKK182 pKa = 10.41IDD184 pKa = 3.44EE185 pKa = 4.88WDD187 pKa = 3.29NSTYY191 pKa = 8.59QTNTYY196 pKa = 10.17FGFHH200 pKa = 7.3AISGGRR206 pKa = 11.84LDD208 pKa = 5.07DD209 pKa = 4.5FSVTDD214 pKa = 3.71VVTSGISIEE223 pKa = 4.03VHH225 pKa = 5.66GDD227 pKa = 3.49EE228 pKa = 5.07IVDD231 pKa = 4.37AGTEE235 pKa = 4.19DD236 pKa = 3.67TGSVPQGSWVTVPIEE251 pKa = 4.09MIGVSASTVTVNNFAVSGDD270 pKa = 3.94LLFDD274 pKa = 5.09DD275 pKa = 5.46SDD277 pKa = 4.25DD278 pKa = 4.11FSSFTLDD285 pKa = 3.02QDD287 pKa = 3.57EE288 pKa = 4.59TNKK291 pKa = 9.67IRR293 pKa = 11.84VRR295 pKa = 11.84LSAATAGSKK304 pKa = 9.26TGTVTVTYY312 pKa = 8.23NTTEE316 pKa = 3.89NYY318 pKa = 8.41TINVDD323 pKa = 3.19WTVVAKK329 pKa = 10.01TPASDD334 pKa = 3.3VALTKK339 pKa = 10.79GGGGSAPTDD348 pKa = 3.61PTRR351 pKa = 11.84TSYY354 pKa = 11.22KK355 pKa = 9.88PGGMWTQVPFAQTVWADD372 pKa = 3.23EE373 pKa = 4.07TTYY376 pKa = 10.68IGVIAMQEE384 pKa = 4.0GGIEE388 pKa = 5.07SVDD391 pKa = 3.99FWWNGNSVTVNTMTTHH407 pKa = 7.19DD408 pKa = 5.22GISAYY413 pKa = 10.59YY414 pKa = 9.92CAISPNAASGNEE426 pKa = 4.05QIYY429 pKa = 8.52ATINAADD436 pKa = 4.75PYY438 pKa = 10.16AQEE441 pKa = 4.83KK442 pKa = 10.5VLDD445 pKa = 3.99DD446 pKa = 3.93PFYY449 pKa = 10.59FTIFDD454 pKa = 3.73SATVNEE460 pKa = 4.34VHH462 pKa = 6.45VGRR465 pKa = 11.84GDD467 pKa = 3.33ASAGSGTEE475 pKa = 3.69GDD477 pKa = 4.66PYY479 pKa = 11.38LSLQSAINDD488 pKa = 3.43LSLTDD493 pKa = 3.82GGIVTIHH500 pKa = 6.94EE501 pKa = 4.61GTVDD505 pKa = 3.25WPYY508 pKa = 10.49TATPKK513 pKa = 10.65ANSGVFWLRR522 pKa = 11.84MAEE525 pKa = 4.38GEE527 pKa = 4.2DD528 pKa = 3.57RR529 pKa = 11.84YY530 pKa = 11.33NVVKK534 pKa = 10.84NFTGTASTSIRR545 pKa = 11.84LSVEE549 pKa = 3.63RR550 pKa = 11.84LKK552 pKa = 11.39VSGIEE557 pKa = 3.73WDD559 pKa = 3.72TANAAVFYY567 pKa = 10.55NRR569 pKa = 11.84GVSGYY574 pKa = 7.77YY575 pKa = 8.47TVWDD579 pKa = 5.39DD580 pKa = 3.28IHH582 pKa = 8.25VGNSDD587 pKa = 3.73GRR589 pKa = 11.84FTLVNGNHH597 pKa = 6.81LNTGSIGRR605 pKa = 11.84AAGTTDD611 pKa = 3.35KK612 pKa = 11.52DD613 pKa = 3.54GFQNCRR619 pKa = 11.84FVDD622 pKa = 4.67TYY624 pKa = 11.3QGTTGFYY631 pKa = 10.49SINDD635 pKa = 3.28YY636 pKa = 9.98VEE638 pKa = 4.29RR639 pKa = 11.84VLRR642 pKa = 11.84DD643 pKa = 3.49HH644 pKa = 5.66STCWSVANMVSRR656 pKa = 11.84RR657 pKa = 11.84GDD659 pKa = 3.27YY660 pKa = 10.71DD661 pKa = 3.61YY662 pKa = 11.75LRR664 pKa = 11.84DD665 pKa = 3.73NLAALTITYY674 pKa = 10.23SGGATATYY682 pKa = 11.15NLTSTNAGSSRR693 pKa = 11.84DD694 pKa = 3.7LVLLEE699 pKa = 4.81DD700 pKa = 4.72ASPVLTINLKK710 pKa = 8.23TSSSAVVDD718 pKa = 3.53TDD720 pKa = 4.46DD721 pKa = 5.19ADD723 pKa = 4.31GEE725 pKa = 4.34IDD727 pKa = 3.24AHH729 pKa = 6.42TIAQVVSAINATTDD743 pKa = 2.62WSASIDD749 pKa = 3.51TDD751 pKa = 3.87YY752 pKa = 11.56TSAAADD758 pKa = 3.9RR759 pKa = 11.84QLDD762 pKa = 3.71ATYY765 pKa = 10.97LKK767 pKa = 10.21IDD769 pKa = 3.36AHH771 pKa = 6.45FSAVTGGAIPVDD783 pKa = 4.64LYY785 pKa = 11.36VAADD789 pKa = 3.47LHH791 pKa = 7.37VDD793 pKa = 5.27GIQSSYY799 pKa = 8.78GTGVAYY805 pKa = 10.77NRR807 pKa = 11.84FYY809 pKa = 11.45YY810 pKa = 10.82NCDD813 pKa = 3.26FDD815 pKa = 5.84NNPADD820 pKa = 3.63MVATYY825 pKa = 10.89GNNEE829 pKa = 3.97SDD831 pKa = 3.54MQGLFIVGTNGGHH844 pKa = 6.16TDD846 pKa = 3.07GGAYY850 pKa = 9.83FMHH853 pKa = 6.63YY854 pKa = 10.68KK855 pKa = 10.49NVDD858 pKa = 3.18ADD860 pKa = 3.72NASQFIGEE868 pKa = 4.06MDD870 pKa = 4.1NFNMMFCTFEE880 pKa = 4.47GPQIMSPRR888 pKa = 11.84TDD890 pKa = 2.65KK891 pKa = 11.37VGNQEE896 pKa = 4.41YY897 pKa = 10.27IPTNVVFRR905 pKa = 11.84GNAFLHH911 pKa = 6.27LDD913 pKa = 3.06NWEE916 pKa = 4.1DD917 pKa = 3.57QFGGEE922 pKa = 5.25LYY924 pKa = 10.37FANNSQVTDD933 pKa = 3.29DD934 pKa = 3.91WSAIGYY940 pKa = 10.38SNTQSANVAAWDD952 pKa = 4.22LNTDD956 pKa = 3.54GTVGDD961 pKa = 4.47SSTLLDD967 pKa = 3.56RR968 pKa = 11.84SLNGTAYY975 pKa = 8.11QTLWPNGDD983 pKa = 3.28SMASDD988 pKa = 3.84GSDD991 pKa = 4.67PIGAWPPEE999 pKa = 4.42GGVTPTPNGDD1009 pKa = 3.69DD1010 pKa = 4.11DD1011 pKa = 5.45GEE1013 pKa = 4.19EE1014 pKa = 4.08SLLLRR1019 pKa = 11.84FAQRR1023 pKa = 11.84TWRR1026 pKa = 11.84PQWRR1030 pKa = 11.84RR1031 pKa = 11.84DD1032 pKa = 3.32MSLLRR1037 pKa = 11.84SQQQ1040 pKa = 2.93
MM1 pKa = 7.31SRR3 pKa = 11.84FAFVFAVLCLTGSFVQAQTTLYY25 pKa = 10.61SDD27 pKa = 3.98SFTNDD32 pKa = 2.28GSAVNLPSWDD42 pKa = 3.52TVEE45 pKa = 5.49NGGVWWNDD53 pKa = 3.1GSDD56 pKa = 3.54STPWVTWSGTDD67 pKa = 3.48SVAHH71 pKa = 6.51HH72 pKa = 6.67SLDD75 pKa = 4.27AIVPGSEE82 pKa = 3.79HH83 pKa = 7.35SGIDD87 pKa = 3.37FGAADD92 pKa = 3.67VDD94 pKa = 3.58ITFEE98 pKa = 4.21FNYY101 pKa = 9.12GTGTTASYY109 pKa = 11.51DD110 pKa = 3.69SIFWIRR116 pKa = 11.84ANSDD120 pKa = 2.77RR121 pKa = 11.84SEE123 pKa = 3.99ALGIYY128 pKa = 10.03HH129 pKa = 6.54SVVDD133 pKa = 5.33DD134 pKa = 4.03EE135 pKa = 4.68FTLVDD140 pKa = 4.12EE141 pKa = 5.38DD142 pKa = 4.36EE143 pKa = 4.56VDD145 pKa = 3.8LGEE148 pKa = 4.34TPVASGLTTANTYY161 pKa = 7.02YY162 pKa = 10.38TIRR165 pKa = 11.84ILLSGSKK172 pKa = 9.94ISLWVDD178 pKa = 3.18DD179 pKa = 4.15VLKK182 pKa = 10.41IDD184 pKa = 3.44EE185 pKa = 4.88WDD187 pKa = 3.29NSTYY191 pKa = 8.59QTNTYY196 pKa = 10.17FGFHH200 pKa = 7.3AISGGRR206 pKa = 11.84LDD208 pKa = 5.07DD209 pKa = 4.5FSVTDD214 pKa = 3.71VVTSGISIEE223 pKa = 4.03VHH225 pKa = 5.66GDD227 pKa = 3.49EE228 pKa = 5.07IVDD231 pKa = 4.37AGTEE235 pKa = 4.19DD236 pKa = 3.67TGSVPQGSWVTVPIEE251 pKa = 4.09MIGVSASTVTVNNFAVSGDD270 pKa = 3.94LLFDD274 pKa = 5.09DD275 pKa = 5.46SDD277 pKa = 4.25DD278 pKa = 4.11FSSFTLDD285 pKa = 3.02QDD287 pKa = 3.57EE288 pKa = 4.59TNKK291 pKa = 9.67IRR293 pKa = 11.84VRR295 pKa = 11.84LSAATAGSKK304 pKa = 9.26TGTVTVTYY312 pKa = 8.23NTTEE316 pKa = 3.89NYY318 pKa = 8.41TINVDD323 pKa = 3.19WTVVAKK329 pKa = 10.01TPASDD334 pKa = 3.3VALTKK339 pKa = 10.79GGGGSAPTDD348 pKa = 3.61PTRR351 pKa = 11.84TSYY354 pKa = 11.22KK355 pKa = 9.88PGGMWTQVPFAQTVWADD372 pKa = 3.23EE373 pKa = 4.07TTYY376 pKa = 10.68IGVIAMQEE384 pKa = 4.0GGIEE388 pKa = 5.07SVDD391 pKa = 3.99FWWNGNSVTVNTMTTHH407 pKa = 7.19DD408 pKa = 5.22GISAYY413 pKa = 10.59YY414 pKa = 9.92CAISPNAASGNEE426 pKa = 4.05QIYY429 pKa = 8.52ATINAADD436 pKa = 4.75PYY438 pKa = 10.16AQEE441 pKa = 4.83KK442 pKa = 10.5VLDD445 pKa = 3.99DD446 pKa = 3.93PFYY449 pKa = 10.59FTIFDD454 pKa = 3.73SATVNEE460 pKa = 4.34VHH462 pKa = 6.45VGRR465 pKa = 11.84GDD467 pKa = 3.33ASAGSGTEE475 pKa = 3.69GDD477 pKa = 4.66PYY479 pKa = 11.38LSLQSAINDD488 pKa = 3.43LSLTDD493 pKa = 3.82GGIVTIHH500 pKa = 6.94EE501 pKa = 4.61GTVDD505 pKa = 3.25WPYY508 pKa = 10.49TATPKK513 pKa = 10.65ANSGVFWLRR522 pKa = 11.84MAEE525 pKa = 4.38GEE527 pKa = 4.2DD528 pKa = 3.57RR529 pKa = 11.84YY530 pKa = 11.33NVVKK534 pKa = 10.84NFTGTASTSIRR545 pKa = 11.84LSVEE549 pKa = 3.63RR550 pKa = 11.84LKK552 pKa = 11.39VSGIEE557 pKa = 3.73WDD559 pKa = 3.72TANAAVFYY567 pKa = 10.55NRR569 pKa = 11.84GVSGYY574 pKa = 7.77YY575 pKa = 8.47TVWDD579 pKa = 5.39DD580 pKa = 3.28IHH582 pKa = 8.25VGNSDD587 pKa = 3.73GRR589 pKa = 11.84FTLVNGNHH597 pKa = 6.81LNTGSIGRR605 pKa = 11.84AAGTTDD611 pKa = 3.35KK612 pKa = 11.52DD613 pKa = 3.54GFQNCRR619 pKa = 11.84FVDD622 pKa = 4.67TYY624 pKa = 11.3QGTTGFYY631 pKa = 10.49SINDD635 pKa = 3.28YY636 pKa = 9.98VEE638 pKa = 4.29RR639 pKa = 11.84VLRR642 pKa = 11.84DD643 pKa = 3.49HH644 pKa = 5.66STCWSVANMVSRR656 pKa = 11.84RR657 pKa = 11.84GDD659 pKa = 3.27YY660 pKa = 10.71DD661 pKa = 3.61YY662 pKa = 11.75LRR664 pKa = 11.84DD665 pKa = 3.73NLAALTITYY674 pKa = 10.23SGGATATYY682 pKa = 11.15NLTSTNAGSSRR693 pKa = 11.84DD694 pKa = 3.7LVLLEE699 pKa = 4.81DD700 pKa = 4.72ASPVLTINLKK710 pKa = 8.23TSSSAVVDD718 pKa = 3.53TDD720 pKa = 4.46DD721 pKa = 5.19ADD723 pKa = 4.31GEE725 pKa = 4.34IDD727 pKa = 3.24AHH729 pKa = 6.42TIAQVVSAINATTDD743 pKa = 2.62WSASIDD749 pKa = 3.51TDD751 pKa = 3.87YY752 pKa = 11.56TSAAADD758 pKa = 3.9RR759 pKa = 11.84QLDD762 pKa = 3.71ATYY765 pKa = 10.97LKK767 pKa = 10.21IDD769 pKa = 3.36AHH771 pKa = 6.45FSAVTGGAIPVDD783 pKa = 4.64LYY785 pKa = 11.36VAADD789 pKa = 3.47LHH791 pKa = 7.37VDD793 pKa = 5.27GIQSSYY799 pKa = 8.78GTGVAYY805 pKa = 10.77NRR807 pKa = 11.84FYY809 pKa = 11.45YY810 pKa = 10.82NCDD813 pKa = 3.26FDD815 pKa = 5.84NNPADD820 pKa = 3.63MVATYY825 pKa = 10.89GNNEE829 pKa = 3.97SDD831 pKa = 3.54MQGLFIVGTNGGHH844 pKa = 6.16TDD846 pKa = 3.07GGAYY850 pKa = 9.83FMHH853 pKa = 6.63YY854 pKa = 10.68KK855 pKa = 10.49NVDD858 pKa = 3.18ADD860 pKa = 3.72NASQFIGEE868 pKa = 4.06MDD870 pKa = 4.1NFNMMFCTFEE880 pKa = 4.47GPQIMSPRR888 pKa = 11.84TDD890 pKa = 2.65KK891 pKa = 11.37VGNQEE896 pKa = 4.41YY897 pKa = 10.27IPTNVVFRR905 pKa = 11.84GNAFLHH911 pKa = 6.27LDD913 pKa = 3.06NWEE916 pKa = 4.1DD917 pKa = 3.57QFGGEE922 pKa = 5.25LYY924 pKa = 10.37FANNSQVTDD933 pKa = 3.29DD934 pKa = 3.91WSAIGYY940 pKa = 10.38SNTQSANVAAWDD952 pKa = 4.22LNTDD956 pKa = 3.54GTVGDD961 pKa = 4.47SSTLLDD967 pKa = 3.56RR968 pKa = 11.84SLNGTAYY975 pKa = 8.11QTLWPNGDD983 pKa = 3.28SMASDD988 pKa = 3.84GSDD991 pKa = 4.67PIGAWPPEE999 pKa = 4.42GGVTPTPNGDD1009 pKa = 3.69DD1010 pKa = 4.11DD1011 pKa = 5.45GEE1013 pKa = 4.19EE1014 pKa = 4.08SLLLRR1019 pKa = 11.84FAQRR1023 pKa = 11.84TWRR1026 pKa = 11.84PQWRR1030 pKa = 11.84RR1031 pKa = 11.84DD1032 pKa = 3.32MSLLRR1037 pKa = 11.84SQQQ1040 pKa = 2.93
Molecular weight: 112.55 kDa
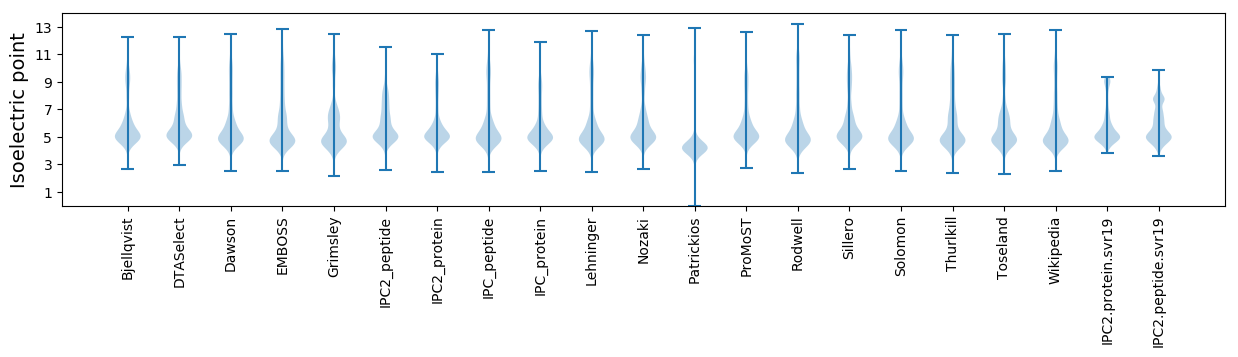
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5C5WPP0|A0A5C5WPP0_9PLAN Xylose isomerase-like TIM barrel OS=Thalassoglobus neptunius OX=1938619 GN=KOR42_30930 PE=4 SV=1
MM1 pKa = 7.27PRR3 pKa = 11.84KK4 pKa = 9.56RR5 pKa = 11.84SQVTIPEE12 pKa = 4.05EE13 pKa = 3.82LATTFATSEE22 pKa = 4.09RR23 pKa = 11.84CPSRR27 pKa = 11.84EE28 pKa = 3.88SVRR31 pKa = 11.84TIDD34 pKa = 3.98LTGTLKK40 pKa = 10.21TIRR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84EE48 pKa = 4.13NCPQSPGVYY57 pKa = 10.24SLINSEE63 pKa = 3.99NQIVYY68 pKa = 10.55VGMSVSLANRR78 pKa = 11.84LQTYY82 pKa = 7.48FTQKK86 pKa = 8.63RR87 pKa = 11.84ARR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 3.8SRR94 pKa = 11.84IRR96 pKa = 11.84RR97 pKa = 11.84AAVRR101 pKa = 11.84LIWQPLDD108 pKa = 3.16HH109 pKa = 6.8GLIARR114 pKa = 11.84IRR116 pKa = 11.84EE117 pKa = 3.85RR118 pKa = 11.84EE119 pKa = 4.35LIRR122 pKa = 11.84KK123 pKa = 7.68FRR125 pKa = 11.84PLMNVIGHH133 pKa = 5.84PTQLKK138 pKa = 7.39TGYY141 pKa = 9.64VVAIDD146 pKa = 3.93HH147 pKa = 6.49PVGGFEE153 pKa = 4.58FVEE156 pKa = 4.42EE157 pKa = 4.15LRR159 pKa = 11.84GRR161 pKa = 11.84YY162 pKa = 8.73QGIWGPIPFNQWSRR176 pKa = 11.84DD177 pKa = 3.36ATEE180 pKa = 3.85EE181 pKa = 4.41LNLHH185 pKa = 5.88FQLRR189 pKa = 11.84DD190 pKa = 3.71CPKK193 pKa = 9.06STVLYY198 pKa = 10.11FSGQPTPEE206 pKa = 4.44APSTPCLRR214 pKa = 11.84ADD216 pKa = 4.84LGTCLSPCIGKK227 pKa = 10.02CSKK230 pKa = 9.24TAYY233 pKa = 10.02LRR235 pKa = 11.84AFKK238 pKa = 10.47RR239 pKa = 11.84ARR241 pKa = 11.84SFLNGSAEE249 pKa = 4.08DD250 pKa = 3.54VTAEE254 pKa = 4.31IEE256 pKa = 4.31TQMRR260 pKa = 11.84EE261 pKa = 3.8AAGEE265 pKa = 4.3KK266 pKa = 10.36NFEE269 pKa = 3.75TAARR273 pKa = 11.84LRR275 pKa = 11.84DD276 pKa = 3.31RR277 pKa = 11.84MKK279 pKa = 10.92AFRR282 pKa = 11.84YY283 pKa = 10.45LNDD286 pKa = 3.47RR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84FHH292 pKa = 7.51DD293 pKa = 2.99WTNRR297 pKa = 11.84ANFVYY302 pKa = 10.38RR303 pKa = 11.84FTSEE307 pKa = 3.88IDD309 pKa = 3.45SQPRR313 pKa = 11.84WLVVSRR319 pKa = 11.84GVIHH323 pKa = 6.51QLIDD327 pKa = 3.53EE328 pKa = 4.74PSSVEE333 pKa = 3.67QAMGILNPLNSSRR346 pKa = 11.84TRR348 pKa = 11.84HH349 pKa = 5.64SKK351 pKa = 7.97PTRR354 pKa = 11.84PNEE357 pKa = 3.75QLRR360 pKa = 11.84PGDD363 pKa = 3.88FEE365 pKa = 3.94TARR368 pKa = 11.84ILFRR372 pKa = 11.84WFRR375 pKa = 11.84QFPKK379 pKa = 10.27EE380 pKa = 3.75KK381 pKa = 9.57TSRR384 pKa = 11.84LSYY387 pKa = 10.38SAALRR392 pKa = 11.84LIKK395 pKa = 10.07RR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84ADD400 pKa = 2.96
MM1 pKa = 7.27PRR3 pKa = 11.84KK4 pKa = 9.56RR5 pKa = 11.84SQVTIPEE12 pKa = 4.05EE13 pKa = 3.82LATTFATSEE22 pKa = 4.09RR23 pKa = 11.84CPSRR27 pKa = 11.84EE28 pKa = 3.88SVRR31 pKa = 11.84TIDD34 pKa = 3.98LTGTLKK40 pKa = 10.21TIRR43 pKa = 11.84SRR45 pKa = 11.84VRR47 pKa = 11.84EE48 pKa = 4.13NCPQSPGVYY57 pKa = 10.24SLINSEE63 pKa = 3.99NQIVYY68 pKa = 10.55VGMSVSLANRR78 pKa = 11.84LQTYY82 pKa = 7.48FTQKK86 pKa = 8.63RR87 pKa = 11.84ARR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84EE92 pKa = 3.8SRR94 pKa = 11.84IRR96 pKa = 11.84RR97 pKa = 11.84AAVRR101 pKa = 11.84LIWQPLDD108 pKa = 3.16HH109 pKa = 6.8GLIARR114 pKa = 11.84IRR116 pKa = 11.84EE117 pKa = 3.85RR118 pKa = 11.84EE119 pKa = 4.35LIRR122 pKa = 11.84KK123 pKa = 7.68FRR125 pKa = 11.84PLMNVIGHH133 pKa = 5.84PTQLKK138 pKa = 7.39TGYY141 pKa = 9.64VVAIDD146 pKa = 3.93HH147 pKa = 6.49PVGGFEE153 pKa = 4.58FVEE156 pKa = 4.42EE157 pKa = 4.15LRR159 pKa = 11.84GRR161 pKa = 11.84YY162 pKa = 8.73QGIWGPIPFNQWSRR176 pKa = 11.84DD177 pKa = 3.36ATEE180 pKa = 3.85EE181 pKa = 4.41LNLHH185 pKa = 5.88FQLRR189 pKa = 11.84DD190 pKa = 3.71CPKK193 pKa = 9.06STVLYY198 pKa = 10.11FSGQPTPEE206 pKa = 4.44APSTPCLRR214 pKa = 11.84ADD216 pKa = 4.84LGTCLSPCIGKK227 pKa = 10.02CSKK230 pKa = 9.24TAYY233 pKa = 10.02LRR235 pKa = 11.84AFKK238 pKa = 10.47RR239 pKa = 11.84ARR241 pKa = 11.84SFLNGSAEE249 pKa = 4.08DD250 pKa = 3.54VTAEE254 pKa = 4.31IEE256 pKa = 4.31TQMRR260 pKa = 11.84EE261 pKa = 3.8AAGEE265 pKa = 4.3KK266 pKa = 10.36NFEE269 pKa = 3.75TAARR273 pKa = 11.84LRR275 pKa = 11.84DD276 pKa = 3.31RR277 pKa = 11.84MKK279 pKa = 10.92AFRR282 pKa = 11.84YY283 pKa = 10.45LNDD286 pKa = 3.47RR287 pKa = 11.84LRR289 pKa = 11.84RR290 pKa = 11.84FHH292 pKa = 7.51DD293 pKa = 2.99WTNRR297 pKa = 11.84ANFVYY302 pKa = 10.38RR303 pKa = 11.84FTSEE307 pKa = 3.88IDD309 pKa = 3.45SQPRR313 pKa = 11.84WLVVSRR319 pKa = 11.84GVIHH323 pKa = 6.51QLIDD327 pKa = 3.53EE328 pKa = 4.74PSSVEE333 pKa = 3.67QAMGILNPLNSSRR346 pKa = 11.84TRR348 pKa = 11.84HH349 pKa = 5.64SKK351 pKa = 7.97PTRR354 pKa = 11.84PNEE357 pKa = 3.75QLRR360 pKa = 11.84PGDD363 pKa = 3.88FEE365 pKa = 3.94TARR368 pKa = 11.84ILFRR372 pKa = 11.84WFRR375 pKa = 11.84QFPKK379 pKa = 10.27EE380 pKa = 3.75KK381 pKa = 9.57TSRR384 pKa = 11.84LSYY387 pKa = 10.38SAALRR392 pKa = 11.84LIKK395 pKa = 10.07RR396 pKa = 11.84RR397 pKa = 11.84RR398 pKa = 11.84ADD400 pKa = 2.96
Molecular weight: 46.37 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1904778 |
29 |
3664 |
348.9 |
38.74 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.109 ± 0.029 | 1.173 ± 0.012 |
6.041 ± 0.028 | 7.109 ± 0.033 |
4.068 ± 0.019 | 7.316 ± 0.038 |
2.235 ± 0.016 | 5.39 ± 0.021 |
3.8 ± 0.029 | 9.672 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.152 ± 0.014 | 3.508 ± 0.025 |
5.069 ± 0.021 | 4.205 ± 0.022 |
6.396 ± 0.028 | 7.192 ± 0.032 |
5.529 ± 0.026 | 7.15 ± 0.027 |
1.509 ± 0.013 | 2.377 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
