
Fer-de-Lance paramyxovirus (strain ATCC VR-895) (FLDV) (Fer-de-Lance virus)
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Orthoparamyxovirinae; Ferlavirus; Reptilian ferlavirus
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
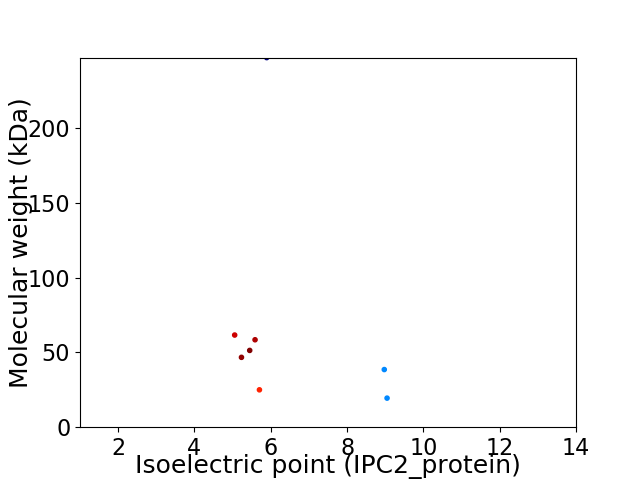
Virtual 2D-PAGE plot for 8 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q6YIS1|Q6YIS1_FDLV8 Fusion glycoprotein F0 OS=Fer-de-Lance paramyxovirus (strain ATCC VR-895) OX=1283347 GN=F PE=3 SV=1
MM1 pKa = 7.93DD2 pKa = 5.49PKK4 pKa = 10.77SYY6 pKa = 11.08YY7 pKa = 10.54CNEE10 pKa = 4.36DD11 pKa = 3.18LRR13 pKa = 11.84SDD15 pKa = 3.79GGEE18 pKa = 3.92KK19 pKa = 10.59SPGGDD24 pKa = 3.17LYY26 pKa = 11.32KK27 pKa = 11.02GIILVSTVISLIIAIISLAFIIDD50 pKa = 3.52NKK52 pKa = 10.76INIQSLDD59 pKa = 3.73PLRR62 pKa = 11.84GLEE65 pKa = 4.11DD66 pKa = 3.63SYY68 pKa = 11.85LVPIKK73 pKa = 10.59DD74 pKa = 3.36KK75 pKa = 11.41SEE77 pKa = 4.72SISQDD82 pKa = 2.64IQEE85 pKa = 5.07GIFPRR90 pKa = 11.84LNLITAATTTTIPRR104 pKa = 11.84SIAIQTKK111 pKa = 10.09DD112 pKa = 3.61LSDD115 pKa = 5.41LIMNRR120 pKa = 11.84CYY122 pKa = 10.48PSVVNNDD129 pKa = 3.33TSCDD133 pKa = 3.57VLAGAIHH140 pKa = 6.76SNLFSQLDD148 pKa = 3.65PSTYY152 pKa = 6.92WTCSSGTPTMNQTVKK167 pKa = 10.92LLPDD171 pKa = 3.63NSQIPGSTYY180 pKa = 9.1STGCVRR186 pKa = 11.84IPTFSLGSMIYY197 pKa = 9.69SYY199 pKa = 10.73SHH201 pKa = 5.07NVIYY205 pKa = 9.98EE206 pKa = 3.91GCNDD210 pKa = 3.42HH211 pKa = 7.04SKK213 pKa = 10.93SSQYY217 pKa = 8.87WQLGYY222 pKa = 10.41ISTSKK227 pKa = 9.57TGEE230 pKa = 3.85PLQQVSRR237 pKa = 11.84TLTLNNGLNRR247 pKa = 11.84KK248 pKa = 8.96SCSTVAQGRR257 pKa = 11.84GAYY260 pKa = 9.35LLCTNVVEE268 pKa = 5.87DD269 pKa = 4.05EE270 pKa = 4.2RR271 pKa = 11.84TDD273 pKa = 3.48YY274 pKa = 10.32STEE277 pKa = 4.35GIQDD281 pKa = 3.51LTLDD285 pKa = 4.05YY286 pKa = 10.95IDD288 pKa = 3.94IFGAEE293 pKa = 3.34RR294 pKa = 11.84SYY296 pKa = 11.1RR297 pKa = 11.84YY298 pKa = 8.44TNNEE302 pKa = 3.16VDD304 pKa = 3.94LDD306 pKa = 3.81RR307 pKa = 11.84PYY309 pKa = 11.27AALYY313 pKa = 9.38PSVGSGTVYY322 pKa = 10.77NDD324 pKa = 3.84RR325 pKa = 11.84ILFLGYY331 pKa = 10.09GGLMTPYY338 pKa = 10.36GDD340 pKa = 4.0QAMCQAPEE348 pKa = 4.39CTSATQEE355 pKa = 4.28GCNSNQLIGYY365 pKa = 8.23FSGRR369 pKa = 11.84QIVNCIIEE377 pKa = 4.81IITVGTEE384 pKa = 3.54KK385 pKa = 10.65PIIRR389 pKa = 11.84VRR391 pKa = 11.84TIPNSQVWLGAEE403 pKa = 4.28GRR405 pKa = 11.84IQTLGGVLYY414 pKa = 10.43LYY416 pKa = 10.08IRR418 pKa = 11.84SSGWHH423 pKa = 6.36ALAQTGIILTLDD435 pKa = 4.84PIRR438 pKa = 11.84ISWIVNTGYY447 pKa = 10.51SRR449 pKa = 11.84PGNGPCSASSRR460 pKa = 11.84CPAQCITGVYY470 pKa = 9.63TDD472 pKa = 4.94IFPLSQNYY480 pKa = 9.96GYY482 pKa = 10.66LATVTLLSGVDD493 pKa = 3.17RR494 pKa = 11.84VNPVISYY501 pKa = 7.4GTSTGRR507 pKa = 11.84VADD510 pKa = 3.87SQLTSSSQVAAYY522 pKa = 7.11TTTTCFTFNQKK533 pKa = 10.0GYY535 pKa = 9.02CYY537 pKa = 10.0HH538 pKa = 7.93IIEE541 pKa = 5.32LSPATLGIFQPVLVVTEE558 pKa = 4.09IPKK561 pKa = 9.79ICSS564 pKa = 3.11
MM1 pKa = 7.93DD2 pKa = 5.49PKK4 pKa = 10.77SYY6 pKa = 11.08YY7 pKa = 10.54CNEE10 pKa = 4.36DD11 pKa = 3.18LRR13 pKa = 11.84SDD15 pKa = 3.79GGEE18 pKa = 3.92KK19 pKa = 10.59SPGGDD24 pKa = 3.17LYY26 pKa = 11.32KK27 pKa = 11.02GIILVSTVISLIIAIISLAFIIDD50 pKa = 3.52NKK52 pKa = 10.76INIQSLDD59 pKa = 3.73PLRR62 pKa = 11.84GLEE65 pKa = 4.11DD66 pKa = 3.63SYY68 pKa = 11.85LVPIKK73 pKa = 10.59DD74 pKa = 3.36KK75 pKa = 11.41SEE77 pKa = 4.72SISQDD82 pKa = 2.64IQEE85 pKa = 5.07GIFPRR90 pKa = 11.84LNLITAATTTTIPRR104 pKa = 11.84SIAIQTKK111 pKa = 10.09DD112 pKa = 3.61LSDD115 pKa = 5.41LIMNRR120 pKa = 11.84CYY122 pKa = 10.48PSVVNNDD129 pKa = 3.33TSCDD133 pKa = 3.57VLAGAIHH140 pKa = 6.76SNLFSQLDD148 pKa = 3.65PSTYY152 pKa = 6.92WTCSSGTPTMNQTVKK167 pKa = 10.92LLPDD171 pKa = 3.63NSQIPGSTYY180 pKa = 9.1STGCVRR186 pKa = 11.84IPTFSLGSMIYY197 pKa = 9.69SYY199 pKa = 10.73SHH201 pKa = 5.07NVIYY205 pKa = 9.98EE206 pKa = 3.91GCNDD210 pKa = 3.42HH211 pKa = 7.04SKK213 pKa = 10.93SSQYY217 pKa = 8.87WQLGYY222 pKa = 10.41ISTSKK227 pKa = 9.57TGEE230 pKa = 3.85PLQQVSRR237 pKa = 11.84TLTLNNGLNRR247 pKa = 11.84KK248 pKa = 8.96SCSTVAQGRR257 pKa = 11.84GAYY260 pKa = 9.35LLCTNVVEE268 pKa = 5.87DD269 pKa = 4.05EE270 pKa = 4.2RR271 pKa = 11.84TDD273 pKa = 3.48YY274 pKa = 10.32STEE277 pKa = 4.35GIQDD281 pKa = 3.51LTLDD285 pKa = 4.05YY286 pKa = 10.95IDD288 pKa = 3.94IFGAEE293 pKa = 3.34RR294 pKa = 11.84SYY296 pKa = 11.1RR297 pKa = 11.84YY298 pKa = 8.44TNNEE302 pKa = 3.16VDD304 pKa = 3.94LDD306 pKa = 3.81RR307 pKa = 11.84PYY309 pKa = 11.27AALYY313 pKa = 9.38PSVGSGTVYY322 pKa = 10.77NDD324 pKa = 3.84RR325 pKa = 11.84ILFLGYY331 pKa = 10.09GGLMTPYY338 pKa = 10.36GDD340 pKa = 4.0QAMCQAPEE348 pKa = 4.39CTSATQEE355 pKa = 4.28GCNSNQLIGYY365 pKa = 8.23FSGRR369 pKa = 11.84QIVNCIIEE377 pKa = 4.81IITVGTEE384 pKa = 3.54KK385 pKa = 10.65PIIRR389 pKa = 11.84VRR391 pKa = 11.84TIPNSQVWLGAEE403 pKa = 4.28GRR405 pKa = 11.84IQTLGGVLYY414 pKa = 10.43LYY416 pKa = 10.08IRR418 pKa = 11.84SSGWHH423 pKa = 6.36ALAQTGIILTLDD435 pKa = 4.84PIRR438 pKa = 11.84ISWIVNTGYY447 pKa = 10.51SRR449 pKa = 11.84PGNGPCSASSRR460 pKa = 11.84CPAQCITGVYY470 pKa = 9.63TDD472 pKa = 4.94IFPLSQNYY480 pKa = 9.96GYY482 pKa = 10.66LATVTLLSGVDD493 pKa = 3.17RR494 pKa = 11.84VNPVISYY501 pKa = 7.4GTSTGRR507 pKa = 11.84VADD510 pKa = 3.87SQLTSSSQVAAYY522 pKa = 7.11TTTTCFTFNQKK533 pKa = 10.0GYY535 pKa = 9.02CYY537 pKa = 10.0HH538 pKa = 7.93IIEE541 pKa = 5.32LSPATLGIFQPVLVVTEE558 pKa = 4.09IPKK561 pKa = 9.79ICSS564 pKa = 3.11
Molecular weight: 61.6 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q6YIS6|Q6YIS6_FDLV8 Nucleocapsid OS=Fer-de-Lance paramyxovirus (strain ATCC VR-895) OX=1283347 GN=NP PE=3 SV=1
MM1 pKa = 7.27IRR3 pKa = 11.84TRR5 pKa = 11.84IYY7 pKa = 10.39KK8 pKa = 8.35PTYY11 pKa = 6.94TTTTPPTCHH20 pKa = 6.15TPIKK24 pKa = 9.83MEE26 pKa = 4.15EE27 pKa = 4.0DD28 pKa = 3.7PRR30 pKa = 11.84EE31 pKa = 4.05KK32 pKa = 9.72MHH34 pKa = 6.32PQSMWRR40 pKa = 11.84LVRR43 pKa = 11.84LRR45 pKa = 11.84AQRR48 pKa = 11.84LLSYY52 pKa = 10.72SEE54 pKa = 4.16STDD57 pKa = 3.08LSTRR61 pKa = 11.84EE62 pKa = 3.85FLEE65 pKa = 4.51DD66 pKa = 3.24VSKK69 pKa = 11.25SVVVLFNRR77 pKa = 11.84DD78 pKa = 3.08GMSSISQWRR87 pKa = 11.84TEE89 pKa = 3.85DD90 pKa = 3.29CAARR94 pKa = 11.84RR95 pKa = 11.84LGNLSKK101 pKa = 10.44FAWDD105 pKa = 3.34AVTKK109 pKa = 10.83GRR111 pKa = 11.84MDD113 pKa = 3.86PCRR116 pKa = 11.84LAFKK120 pKa = 10.18MVTEE124 pKa = 5.03LGNDD128 pKa = 3.15VAIRR132 pKa = 11.84AEE134 pKa = 3.81ILTVVWLITGWSTIPRR150 pKa = 11.84TLHH153 pKa = 5.97KK154 pKa = 10.29DD155 pKa = 2.99LWSSAIYY162 pKa = 10.06RR163 pKa = 11.84RR164 pKa = 11.84LSLL167 pKa = 4.06
MM1 pKa = 7.27IRR3 pKa = 11.84TRR5 pKa = 11.84IYY7 pKa = 10.39KK8 pKa = 8.35PTYY11 pKa = 6.94TTTTPPTCHH20 pKa = 6.15TPIKK24 pKa = 9.83MEE26 pKa = 4.15EE27 pKa = 4.0DD28 pKa = 3.7PRR30 pKa = 11.84EE31 pKa = 4.05KK32 pKa = 9.72MHH34 pKa = 6.32PQSMWRR40 pKa = 11.84LVRR43 pKa = 11.84LRR45 pKa = 11.84AQRR48 pKa = 11.84LLSYY52 pKa = 10.72SEE54 pKa = 4.16STDD57 pKa = 3.08LSTRR61 pKa = 11.84EE62 pKa = 3.85FLEE65 pKa = 4.51DD66 pKa = 3.24VSKK69 pKa = 11.25SVVVLFNRR77 pKa = 11.84DD78 pKa = 3.08GMSSISQWRR87 pKa = 11.84TEE89 pKa = 3.85DD90 pKa = 3.29CAARR94 pKa = 11.84RR95 pKa = 11.84LGNLSKK101 pKa = 10.44FAWDD105 pKa = 3.34AVTKK109 pKa = 10.83GRR111 pKa = 11.84MDD113 pKa = 3.86PCRR116 pKa = 11.84LAFKK120 pKa = 10.18MVTEE124 pKa = 5.03LGNDD128 pKa = 3.15VAIRR132 pKa = 11.84AEE134 pKa = 3.81ILTVVWLITGWSTIPRR150 pKa = 11.84TLHH153 pKa = 5.97KK154 pKa = 10.29DD155 pKa = 2.99LWSSAIYY162 pKa = 10.06RR163 pKa = 11.84RR164 pKa = 11.84LSLL167 pKa = 4.06
Molecular weight: 19.44 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4935 |
167 |
2181 |
616.9 |
68.51 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.694 ± 0.775 | 1.743 ± 0.274 |
5.39 ± 0.18 | 5.897 ± 0.639 |
3.019 ± 0.344 | 6.403 ± 0.542 |
1.662 ± 0.33 | 8.186 ± 0.52 |
5.694 ± 0.483 | 9.463 ± 0.784 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.614 ± 0.314 | 4.721 ± 0.313 |
4.093 ± 0.547 | 3.708 ± 0.258 |
4.965 ± 0.488 | 8.936 ± 0.247 |
7.295 ± 0.613 | 6.444 ± 0.354 |
0.993 ± 0.147 | 3.08 ± 0.655 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
