
Eptesicus serotinus papillomavirus 3
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Dyopsipapillomavirus; Dyopsipapillomavirus 1
Average proteome isoelectric point is 6.49
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 6 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
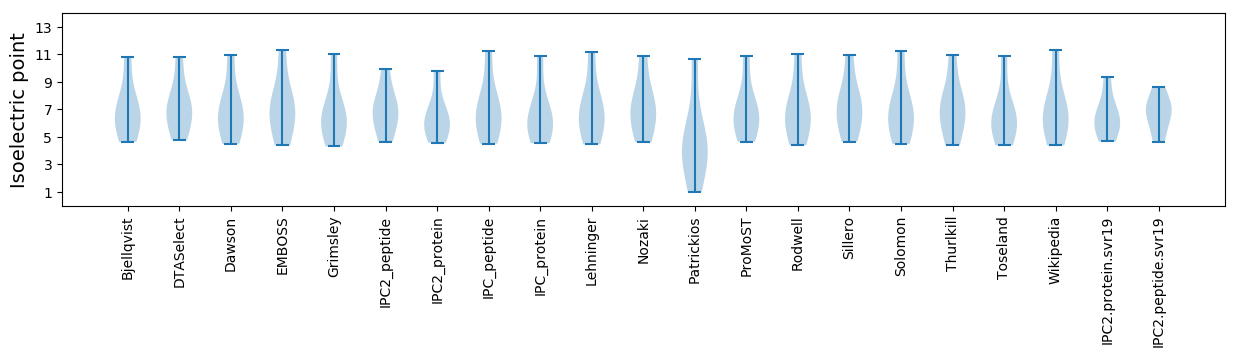
* You can choose from 21 different methods for calculating isoelectric point
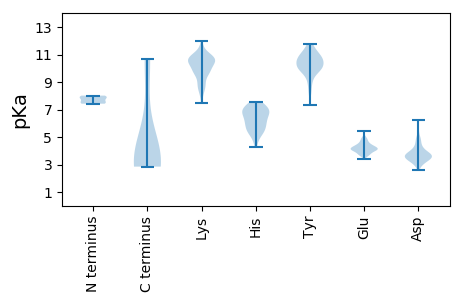
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|W8EG03|W8EG03_9PAPI Regulatory protein E2 OS=Eptesicus serotinus papillomavirus 3 OX=1464073 GN=E2 PE=3 SV=1
MM1 pKa = 7.6ALLLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.81RR10 pKa = 11.84DD11 pKa = 3.32TVEE14 pKa = 3.62NIYY17 pKa = 10.03RR18 pKa = 11.84HH19 pKa = 6.5CIRR22 pKa = 11.84FGTCPPDD29 pKa = 3.36VKK31 pKa = 11.19NKK33 pKa = 9.74VEE35 pKa = 3.96QSTVADD41 pKa = 4.54KK42 pKa = 10.75ILKK45 pKa = 8.34YY46 pKa = 10.87GSGAVYY52 pKa = 10.25FGGLGIGTGSGRR64 pKa = 11.84GSTGVSVGRR73 pKa = 11.84EE74 pKa = 3.72VPIRR78 pKa = 11.84PFRR81 pKa = 11.84PHH83 pKa = 6.9GPIDD87 pKa = 3.61TIGIEE92 pKa = 4.26NIGPRR97 pKa = 11.84EE98 pKa = 4.1TPFAPPDD105 pKa = 3.67RR106 pKa = 11.84APVDD110 pKa = 3.36VDD112 pKa = 3.89LGTGHH117 pKa = 6.49VEE119 pKa = 3.95PVDD122 pKa = 3.72PSIITPIDD130 pKa = 3.98LPTDD134 pKa = 3.44IPTSSVDD141 pKa = 3.32EE142 pKa = 4.91SIVVPTPEE150 pKa = 3.67ITIGGDD156 pKa = 2.93TTAIIEE162 pKa = 4.3VTPNVPDD169 pKa = 3.24NTIISRR175 pKa = 11.84SQYY178 pKa = 10.57NNPAFDD184 pKa = 3.29ISLFNNSTAGEE195 pKa = 4.3LSASDD200 pKa = 4.81HH201 pKa = 5.87ITVSGGGGSIIGGHH215 pKa = 5.97FPLAEE220 pKa = 4.23PEE222 pKa = 4.16GEE224 pKa = 4.17EE225 pKa = 3.73IGLAEE230 pKa = 4.11FTPGVSRR237 pKa = 11.84FRR239 pKa = 11.84PRR241 pKa = 11.84EE242 pKa = 3.77VADD245 pKa = 3.54EE246 pKa = 4.72AEE248 pKa = 4.32TSFITSTPEE257 pKa = 3.37GARR260 pKa = 11.84VGRR263 pKa = 11.84PTRR266 pKa = 11.84VRR268 pKa = 11.84PSLYY272 pKa = 9.7NRR274 pKa = 11.84RR275 pKa = 11.84IEE277 pKa = 4.1QVEE280 pKa = 4.15VTDD283 pKa = 3.9PDD285 pKa = 4.52FIEE288 pKa = 4.57RR289 pKa = 11.84PGRR292 pKa = 11.84LVEE295 pKa = 4.53FGYY298 pKa = 11.05VNPAFEE304 pKa = 4.71GDD306 pKa = 2.87ITEE309 pKa = 4.75IFEE312 pKa = 4.2QDD314 pKa = 3.4LLEE317 pKa = 4.34EE318 pKa = 4.26ARR320 pKa = 11.84AAPSSAFQDD329 pKa = 3.2IVRR332 pKa = 11.84VGRR335 pKa = 11.84QQLSRR340 pKa = 11.84ARR342 pKa = 11.84SGLVRR347 pKa = 11.84VSRR350 pKa = 11.84LGQRR354 pKa = 11.84GTIRR358 pKa = 11.84TRR360 pKa = 11.84SGVRR364 pKa = 11.84IGSTTHH370 pKa = 6.92FYY372 pKa = 10.53MDD374 pKa = 5.21LSPILPDD381 pKa = 3.0EE382 pKa = 4.44SLPIEE387 pKa = 4.36LPGEE391 pKa = 4.02QSMEE395 pKa = 3.77ASIVQPLSEE404 pKa = 4.45EE405 pKa = 4.1GFEE408 pKa = 4.78LVNLDD413 pKa = 3.67EE414 pKa = 4.87VSDD417 pKa = 4.07VVPDD421 pKa = 3.73EE422 pKa = 4.54SLLDD426 pKa = 3.92EE427 pKa = 4.49YY428 pKa = 11.34EE429 pKa = 3.91EE430 pKa = 4.67TEE432 pKa = 4.98FIGSDD437 pKa = 3.48LQLIIHH443 pKa = 7.15DD444 pKa = 4.01EE445 pKa = 4.67DD446 pKa = 3.68IGIEE450 pKa = 4.01RR451 pKa = 11.84DD452 pKa = 3.0SDD454 pKa = 3.67IVVDD458 pKa = 4.23VPSGFIKK465 pKa = 9.81KK466 pKa = 8.43TGTLYY471 pKa = 10.45PDD473 pKa = 3.16IDD475 pKa = 4.04YY476 pKa = 10.26TGVDD480 pKa = 2.75IDD482 pKa = 3.87YY483 pKa = 10.54DD484 pKa = 3.83KK485 pKa = 10.84THH487 pKa = 6.78SGDD490 pKa = 3.51AGDD493 pKa = 5.56IIPADD498 pKa = 3.93TPVITIDD505 pKa = 2.62IWSSDD510 pKa = 3.75FYY512 pKa = 11.21LHH514 pKa = 7.26PSLSRR519 pKa = 11.84KK520 pKa = 8.83RR521 pKa = 11.84KK522 pKa = 8.13RR523 pKa = 11.84RR524 pKa = 11.84RR525 pKa = 11.84KK526 pKa = 9.52FVFVYY531 pKa = 10.67
MM1 pKa = 7.6ALLLRR6 pKa = 11.84RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.81RR10 pKa = 11.84DD11 pKa = 3.32TVEE14 pKa = 3.62NIYY17 pKa = 10.03RR18 pKa = 11.84HH19 pKa = 6.5CIRR22 pKa = 11.84FGTCPPDD29 pKa = 3.36VKK31 pKa = 11.19NKK33 pKa = 9.74VEE35 pKa = 3.96QSTVADD41 pKa = 4.54KK42 pKa = 10.75ILKK45 pKa = 8.34YY46 pKa = 10.87GSGAVYY52 pKa = 10.25FGGLGIGTGSGRR64 pKa = 11.84GSTGVSVGRR73 pKa = 11.84EE74 pKa = 3.72VPIRR78 pKa = 11.84PFRR81 pKa = 11.84PHH83 pKa = 6.9GPIDD87 pKa = 3.61TIGIEE92 pKa = 4.26NIGPRR97 pKa = 11.84EE98 pKa = 4.1TPFAPPDD105 pKa = 3.67RR106 pKa = 11.84APVDD110 pKa = 3.36VDD112 pKa = 3.89LGTGHH117 pKa = 6.49VEE119 pKa = 3.95PVDD122 pKa = 3.72PSIITPIDD130 pKa = 3.98LPTDD134 pKa = 3.44IPTSSVDD141 pKa = 3.32EE142 pKa = 4.91SIVVPTPEE150 pKa = 3.67ITIGGDD156 pKa = 2.93TTAIIEE162 pKa = 4.3VTPNVPDD169 pKa = 3.24NTIISRR175 pKa = 11.84SQYY178 pKa = 10.57NNPAFDD184 pKa = 3.29ISLFNNSTAGEE195 pKa = 4.3LSASDD200 pKa = 4.81HH201 pKa = 5.87ITVSGGGGSIIGGHH215 pKa = 5.97FPLAEE220 pKa = 4.23PEE222 pKa = 4.16GEE224 pKa = 4.17EE225 pKa = 3.73IGLAEE230 pKa = 4.11FTPGVSRR237 pKa = 11.84FRR239 pKa = 11.84PRR241 pKa = 11.84EE242 pKa = 3.77VADD245 pKa = 3.54EE246 pKa = 4.72AEE248 pKa = 4.32TSFITSTPEE257 pKa = 3.37GARR260 pKa = 11.84VGRR263 pKa = 11.84PTRR266 pKa = 11.84VRR268 pKa = 11.84PSLYY272 pKa = 9.7NRR274 pKa = 11.84RR275 pKa = 11.84IEE277 pKa = 4.1QVEE280 pKa = 4.15VTDD283 pKa = 3.9PDD285 pKa = 4.52FIEE288 pKa = 4.57RR289 pKa = 11.84PGRR292 pKa = 11.84LVEE295 pKa = 4.53FGYY298 pKa = 11.05VNPAFEE304 pKa = 4.71GDD306 pKa = 2.87ITEE309 pKa = 4.75IFEE312 pKa = 4.2QDD314 pKa = 3.4LLEE317 pKa = 4.34EE318 pKa = 4.26ARR320 pKa = 11.84AAPSSAFQDD329 pKa = 3.2IVRR332 pKa = 11.84VGRR335 pKa = 11.84QQLSRR340 pKa = 11.84ARR342 pKa = 11.84SGLVRR347 pKa = 11.84VSRR350 pKa = 11.84LGQRR354 pKa = 11.84GTIRR358 pKa = 11.84TRR360 pKa = 11.84SGVRR364 pKa = 11.84IGSTTHH370 pKa = 6.92FYY372 pKa = 10.53MDD374 pKa = 5.21LSPILPDD381 pKa = 3.0EE382 pKa = 4.44SLPIEE387 pKa = 4.36LPGEE391 pKa = 4.02QSMEE395 pKa = 3.77ASIVQPLSEE404 pKa = 4.45EE405 pKa = 4.1GFEE408 pKa = 4.78LVNLDD413 pKa = 3.67EE414 pKa = 4.87VSDD417 pKa = 4.07VVPDD421 pKa = 3.73EE422 pKa = 4.54SLLDD426 pKa = 3.92EE427 pKa = 4.49YY428 pKa = 11.34EE429 pKa = 3.91EE430 pKa = 4.67TEE432 pKa = 4.98FIGSDD437 pKa = 3.48LQLIIHH443 pKa = 7.15DD444 pKa = 4.01EE445 pKa = 4.67DD446 pKa = 3.68IGIEE450 pKa = 4.01RR451 pKa = 11.84DD452 pKa = 3.0SDD454 pKa = 3.67IVVDD458 pKa = 4.23VPSGFIKK465 pKa = 9.81KK466 pKa = 8.43TGTLYY471 pKa = 10.45PDD473 pKa = 3.16IDD475 pKa = 4.04YY476 pKa = 10.26TGVDD480 pKa = 2.75IDD482 pKa = 3.87YY483 pKa = 10.54DD484 pKa = 3.83KK485 pKa = 10.84THH487 pKa = 6.78SGDD490 pKa = 3.51AGDD493 pKa = 5.56IIPADD498 pKa = 3.93TPVITIDD505 pKa = 2.62IWSSDD510 pKa = 3.75FYY512 pKa = 11.21LHH514 pKa = 7.26PSLSRR519 pKa = 11.84KK520 pKa = 8.83RR521 pKa = 11.84KK522 pKa = 8.13RR523 pKa = 11.84RR524 pKa = 11.84RR525 pKa = 11.84KK526 pKa = 9.52FVFVYY531 pKa = 10.67
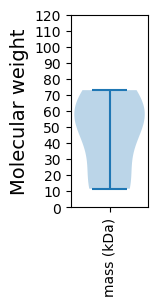
Molecular weight: 58.12 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|W8EH60|W8EH60_9PAPI Replication protein E1 OS=Eptesicus serotinus papillomavirus 3 OX=1464073 GN=E1 PE=3 SV=1
MM1 pKa = 7.92SEE3 pKa = 3.92LQNRR7 pKa = 11.84LDD9 pKa = 4.13YY10 pKa = 11.1IQDD13 pKa = 3.5QILEE17 pKa = 4.31LFEE20 pKa = 4.46KK21 pKa = 10.73QSTDD25 pKa = 3.74LQDD28 pKa = 5.48HH29 pKa = 5.27EE30 pKa = 4.86QFWLLTRR37 pKa = 11.84KK38 pKa = 8.76EE39 pKa = 3.85QAILFLARR47 pKa = 11.84QNNVRR52 pKa = 11.84LSMPVPSLAASKK64 pKa = 10.63ARR66 pKa = 11.84ARR68 pKa = 11.84HH69 pKa = 5.56AIEE72 pKa = 3.78MSLLIKK78 pKa = 10.48SLSEE82 pKa = 3.85SVYY85 pKa = 10.69GRR87 pKa = 11.84EE88 pKa = 3.87AWTLQQVSRR97 pKa = 11.84EE98 pKa = 3.97RR99 pKa = 11.84LLVAPEE105 pKa = 3.77YY106 pKa = 9.86TFKK109 pKa = 11.04KK110 pKa = 9.21GGRR113 pKa = 11.84PVTVMFDD120 pKa = 3.8DD121 pKa = 3.89NHH123 pKa = 7.04DD124 pKa = 3.61NTAEE128 pKa = 3.96YY129 pKa = 8.59TAWDD133 pKa = 3.63SIYY136 pKa = 10.81FQDD139 pKa = 6.02ADD141 pKa = 4.27NEE143 pKa = 4.24WHH145 pKa = 6.74RR146 pKa = 11.84SKK148 pKa = 11.53GDD150 pKa = 2.95VDD152 pKa = 3.88MEE154 pKa = 4.27GLFYY158 pKa = 10.67RR159 pKa = 11.84DD160 pKa = 3.16NDD162 pKa = 3.36GHH164 pKa = 6.96KK165 pKa = 9.81IYY167 pKa = 10.97YY168 pKa = 9.61IDD170 pKa = 3.89FARR173 pKa = 11.84EE174 pKa = 3.42AARR177 pKa = 11.84YY178 pKa = 9.06GSTGKK183 pKa = 9.71WSLLYY188 pKa = 10.85NNNALASVDD197 pKa = 3.63SPGYY201 pKa = 9.29RR202 pKa = 11.84DD203 pKa = 3.34SSGEE207 pKa = 4.02SSTDD211 pKa = 3.23SSPLSARR218 pKa = 11.84SATRR222 pKa = 11.84PTEE225 pKa = 4.39DD226 pKa = 3.09RR227 pKa = 11.84GSPQGSRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84PRR238 pKa = 11.84LWVRR242 pKa = 11.84PRR244 pKa = 11.84SRR246 pKa = 11.84TRR248 pKa = 11.84SRR250 pKa = 11.84SLSRR254 pKa = 11.84SSGRR258 pKa = 11.84SRR260 pKa = 11.84SRR262 pKa = 11.84SRR264 pKa = 11.84SRR266 pKa = 11.84SRR268 pKa = 11.84GATRR272 pKa = 11.84ASIQPAAGTRR282 pKa = 11.84SRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84TRR290 pKa = 11.84SRR292 pKa = 11.84SRR294 pKa = 11.84PRR296 pKa = 11.84SRR298 pKa = 11.84GSPVSPASPASLRR311 pKa = 11.84GGRR314 pKa = 11.84SPGSRR319 pKa = 11.84RR320 pKa = 11.84LGRR323 pKa = 11.84SPRR326 pKa = 11.84PRR328 pKa = 11.84AVDD331 pKa = 3.51PEE333 pKa = 4.25QVGSSKK339 pKa = 10.58RR340 pKa = 11.84SVPAGPKK347 pKa = 9.57RR348 pKa = 11.84RR349 pKa = 11.84LEE351 pKa = 4.09TLLQEE356 pKa = 4.97ARR358 pKa = 11.84DD359 pKa = 4.02PPGLLLTGPPNTLKK373 pKa = 10.7CYY375 pKa = 10.21RR376 pKa = 11.84YY377 pKa = 9.86SLKK380 pKa = 10.59KK381 pKa = 9.71NHH383 pKa = 6.5SSLFSYY389 pKa = 10.6ISTTWHH395 pKa = 4.27WTEE398 pKa = 3.5AKK400 pKa = 8.33GTKK403 pKa = 9.56RR404 pKa = 11.84VGNGKK409 pKa = 8.27MLIMFEE415 pKa = 4.15DD416 pKa = 3.74SEE418 pKa = 4.37RR419 pKa = 11.84RR420 pKa = 11.84EE421 pKa = 4.1WFQKK425 pKa = 9.95IVSLPRR431 pKa = 11.84TLSASKK437 pKa = 10.95VSFAGLL443 pKa = 3.21
MM1 pKa = 7.92SEE3 pKa = 3.92LQNRR7 pKa = 11.84LDD9 pKa = 4.13YY10 pKa = 11.1IQDD13 pKa = 3.5QILEE17 pKa = 4.31LFEE20 pKa = 4.46KK21 pKa = 10.73QSTDD25 pKa = 3.74LQDD28 pKa = 5.48HH29 pKa = 5.27EE30 pKa = 4.86QFWLLTRR37 pKa = 11.84KK38 pKa = 8.76EE39 pKa = 3.85QAILFLARR47 pKa = 11.84QNNVRR52 pKa = 11.84LSMPVPSLAASKK64 pKa = 10.63ARR66 pKa = 11.84ARR68 pKa = 11.84HH69 pKa = 5.56AIEE72 pKa = 3.78MSLLIKK78 pKa = 10.48SLSEE82 pKa = 3.85SVYY85 pKa = 10.69GRR87 pKa = 11.84EE88 pKa = 3.87AWTLQQVSRR97 pKa = 11.84EE98 pKa = 3.97RR99 pKa = 11.84LLVAPEE105 pKa = 3.77YY106 pKa = 9.86TFKK109 pKa = 11.04KK110 pKa = 9.21GGRR113 pKa = 11.84PVTVMFDD120 pKa = 3.8DD121 pKa = 3.89NHH123 pKa = 7.04DD124 pKa = 3.61NTAEE128 pKa = 3.96YY129 pKa = 8.59TAWDD133 pKa = 3.63SIYY136 pKa = 10.81FQDD139 pKa = 6.02ADD141 pKa = 4.27NEE143 pKa = 4.24WHH145 pKa = 6.74RR146 pKa = 11.84SKK148 pKa = 11.53GDD150 pKa = 2.95VDD152 pKa = 3.88MEE154 pKa = 4.27GLFYY158 pKa = 10.67RR159 pKa = 11.84DD160 pKa = 3.16NDD162 pKa = 3.36GHH164 pKa = 6.96KK165 pKa = 9.81IYY167 pKa = 10.97YY168 pKa = 9.61IDD170 pKa = 3.89FARR173 pKa = 11.84EE174 pKa = 3.42AARR177 pKa = 11.84YY178 pKa = 9.06GSTGKK183 pKa = 9.71WSLLYY188 pKa = 10.85NNNALASVDD197 pKa = 3.63SPGYY201 pKa = 9.29RR202 pKa = 11.84DD203 pKa = 3.34SSGEE207 pKa = 4.02SSTDD211 pKa = 3.23SSPLSARR218 pKa = 11.84SATRR222 pKa = 11.84PTEE225 pKa = 4.39DD226 pKa = 3.09RR227 pKa = 11.84GSPQGSRR234 pKa = 11.84RR235 pKa = 11.84RR236 pKa = 11.84PRR238 pKa = 11.84LWVRR242 pKa = 11.84PRR244 pKa = 11.84SRR246 pKa = 11.84TRR248 pKa = 11.84SRR250 pKa = 11.84SLSRR254 pKa = 11.84SSGRR258 pKa = 11.84SRR260 pKa = 11.84SRR262 pKa = 11.84SRR264 pKa = 11.84SRR266 pKa = 11.84SRR268 pKa = 11.84GATRR272 pKa = 11.84ASIQPAAGTRR282 pKa = 11.84SRR284 pKa = 11.84SRR286 pKa = 11.84SRR288 pKa = 11.84TRR290 pKa = 11.84SRR292 pKa = 11.84SRR294 pKa = 11.84PRR296 pKa = 11.84SRR298 pKa = 11.84GSPVSPASPASLRR311 pKa = 11.84GGRR314 pKa = 11.84SPGSRR319 pKa = 11.84RR320 pKa = 11.84LGRR323 pKa = 11.84SPRR326 pKa = 11.84PRR328 pKa = 11.84AVDD331 pKa = 3.51PEE333 pKa = 4.25QVGSSKK339 pKa = 10.58RR340 pKa = 11.84SVPAGPKK347 pKa = 9.57RR348 pKa = 11.84RR349 pKa = 11.84LEE351 pKa = 4.09TLLQEE356 pKa = 4.97ARR358 pKa = 11.84DD359 pKa = 4.02PPGLLLTGPPNTLKK373 pKa = 10.7CYY375 pKa = 10.21RR376 pKa = 11.84YY377 pKa = 9.86SLKK380 pKa = 10.59KK381 pKa = 9.71NHH383 pKa = 6.5SSLFSYY389 pKa = 10.6ISTTWHH395 pKa = 4.27WTEE398 pKa = 3.5AKK400 pKa = 8.33GTKK403 pKa = 9.56RR404 pKa = 11.84VGNGKK409 pKa = 8.27MLIMFEE415 pKa = 4.15DD416 pKa = 3.74SEE418 pKa = 4.37RR419 pKa = 11.84RR420 pKa = 11.84EE421 pKa = 4.1WFQKK425 pKa = 9.95IVSLPRR431 pKa = 11.84TLSASKK437 pKa = 10.95VSFAGLL443 pKa = 3.21
Molecular weight: 50.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2365 |
104 |
642 |
394.2 |
44.42 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.342 ± 0.935 | 1.734 ± 0.729 |
6.554 ± 0.569 | 6.427 ± 0.546 |
4.778 ± 0.743 | 6.3 ± 0.912 |
2.03 ± 0.213 | 5.159 ± 1.197 |
4.524 ± 0.939 | 8.584 ± 0.799 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.86 ± 0.469 | 4.017 ± 0.754 |
5.793 ± 1.032 | 3.975 ± 0.623 |
7.738 ± 1.517 | 8.33 ± 1.477 |
5.539 ± 0.405 | 6.342 ± 0.729 |
1.268 ± 0.342 | 2.706 ± 0.353 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
