
Lactonifactor longoviformis DSM 17459
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Lactonifactor; Lactonifactor longoviformis
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
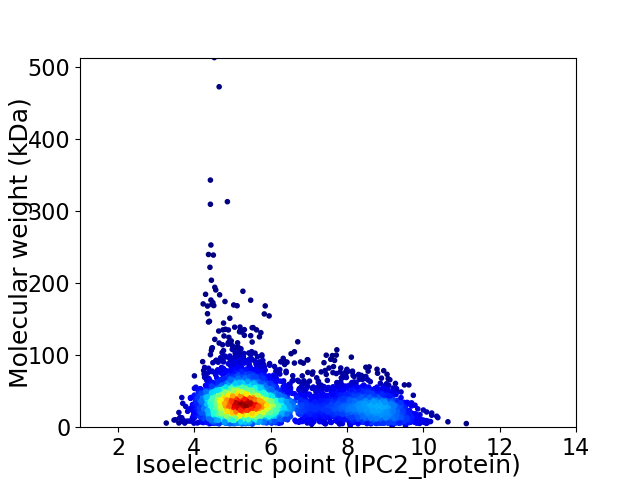
Virtual 2D-PAGE plot for 4380 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1M4SUR5|A0A1M4SUR5_9CLOT Uncharacterized protein OS=Lactonifactor longoviformis DSM 17459 OX=1122155 GN=SAMN02745158_00274 PE=4 SV=1
MM1 pKa = 7.61KK2 pKa = 9.87MNKK5 pKa = 8.15TLSVLCMTVLFAGSLSGCGNDD26 pKa = 3.5GSSPEE31 pKa = 4.22EE32 pKa = 4.15GGSASKK38 pKa = 11.02GGTLNVLTWEE48 pKa = 4.67GDD50 pKa = 3.65VSDD53 pKa = 3.83EE54 pKa = 4.14TVADD58 pKa = 4.14FEE60 pKa = 4.52EE61 pKa = 5.39EE62 pKa = 4.21YY63 pKa = 11.17GVDD66 pKa = 3.57VNITYY71 pKa = 10.09VEE73 pKa = 4.12DD74 pKa = 3.83TNTILAKK81 pKa = 9.55MLQGNSEE88 pKa = 4.11YY89 pKa = 11.09DD90 pKa = 3.59VIDD93 pKa = 3.65IEE95 pKa = 4.39SAYY98 pKa = 10.0MKK100 pKa = 10.57SFVDD104 pKa = 4.33AEE106 pKa = 4.17LLAPLDD112 pKa = 3.61YY113 pKa = 11.43DD114 pKa = 5.13KK115 pKa = 10.66ITNYY119 pKa = 10.65EE120 pKa = 4.2NIDD123 pKa = 3.38PVYY126 pKa = 10.33VEE128 pKa = 4.6KK129 pKa = 10.94GAMGDD134 pKa = 3.5EE135 pKa = 4.25KK136 pKa = 10.59LTYY139 pKa = 9.94SLPITGPLYY148 pKa = 10.44TGVVINKK155 pKa = 6.13EE156 pKa = 4.22TCPIEE161 pKa = 5.16IDD163 pKa = 3.83SFEE166 pKa = 4.82DD167 pKa = 3.38LADD170 pKa = 4.16PALEE174 pKa = 4.17GQVWSTNATISLYY187 pKa = 10.93AGALKK192 pKa = 10.75ALGYY196 pKa = 10.2SPNSQDD202 pKa = 3.23EE203 pKa = 4.47KK204 pKa = 11.1EE205 pKa = 4.26LNEE208 pKa = 4.36AQALLQKK215 pKa = 10.33IKK217 pKa = 10.78PNIKK221 pKa = 10.32AFGASSISSLEE232 pKa = 3.93TGEE235 pKa = 4.66CSVAYY240 pKa = 9.21TYY242 pKa = 10.68DD243 pKa = 3.73YY244 pKa = 10.49NILMFDD250 pKa = 5.55DD251 pKa = 3.82KK252 pKa = 11.92ANWDD256 pKa = 3.47KK257 pKa = 11.61FEE259 pKa = 4.53IVPEE263 pKa = 4.24TTLGYY268 pKa = 8.86CQYY271 pKa = 8.71WTVAASSEE279 pKa = 4.14NQKK282 pKa = 10.27LANEE286 pKa = 5.24FINYY290 pKa = 7.46TYY292 pKa = 10.68SAEE295 pKa = 4.24SAIKK299 pKa = 10.28LLNEE303 pKa = 4.0WGGVPALKK311 pKa = 10.37RR312 pKa = 11.84EE313 pKa = 4.31LIEE316 pKa = 4.17EE317 pKa = 4.61GVAKK321 pKa = 10.39DD322 pKa = 3.61YY323 pKa = 11.4FEE325 pKa = 6.26NPMLQEE331 pKa = 4.39FADD334 pKa = 3.56MWPDD338 pKa = 3.35HH339 pKa = 6.96EE340 pKa = 4.62EE341 pKa = 3.95LSVSDD346 pKa = 4.02EE347 pKa = 3.78QTAIMDD353 pKa = 3.95TLYY356 pKa = 11.04NEE358 pKa = 4.34LMSGEE363 pKa = 4.13
MM1 pKa = 7.61KK2 pKa = 9.87MNKK5 pKa = 8.15TLSVLCMTVLFAGSLSGCGNDD26 pKa = 3.5GSSPEE31 pKa = 4.22EE32 pKa = 4.15GGSASKK38 pKa = 11.02GGTLNVLTWEE48 pKa = 4.67GDD50 pKa = 3.65VSDD53 pKa = 3.83EE54 pKa = 4.14TVADD58 pKa = 4.14FEE60 pKa = 4.52EE61 pKa = 5.39EE62 pKa = 4.21YY63 pKa = 11.17GVDD66 pKa = 3.57VNITYY71 pKa = 10.09VEE73 pKa = 4.12DD74 pKa = 3.83TNTILAKK81 pKa = 9.55MLQGNSEE88 pKa = 4.11YY89 pKa = 11.09DD90 pKa = 3.59VIDD93 pKa = 3.65IEE95 pKa = 4.39SAYY98 pKa = 10.0MKK100 pKa = 10.57SFVDD104 pKa = 4.33AEE106 pKa = 4.17LLAPLDD112 pKa = 3.61YY113 pKa = 11.43DD114 pKa = 5.13KK115 pKa = 10.66ITNYY119 pKa = 10.65EE120 pKa = 4.2NIDD123 pKa = 3.38PVYY126 pKa = 10.33VEE128 pKa = 4.6KK129 pKa = 10.94GAMGDD134 pKa = 3.5EE135 pKa = 4.25KK136 pKa = 10.59LTYY139 pKa = 9.94SLPITGPLYY148 pKa = 10.44TGVVINKK155 pKa = 6.13EE156 pKa = 4.22TCPIEE161 pKa = 5.16IDD163 pKa = 3.83SFEE166 pKa = 4.82DD167 pKa = 3.38LADD170 pKa = 4.16PALEE174 pKa = 4.17GQVWSTNATISLYY187 pKa = 10.93AGALKK192 pKa = 10.75ALGYY196 pKa = 10.2SPNSQDD202 pKa = 3.23EE203 pKa = 4.47KK204 pKa = 11.1EE205 pKa = 4.26LNEE208 pKa = 4.36AQALLQKK215 pKa = 10.33IKK217 pKa = 10.78PNIKK221 pKa = 10.32AFGASSISSLEE232 pKa = 3.93TGEE235 pKa = 4.66CSVAYY240 pKa = 9.21TYY242 pKa = 10.68DD243 pKa = 3.73YY244 pKa = 10.49NILMFDD250 pKa = 5.55DD251 pKa = 3.82KK252 pKa = 11.92ANWDD256 pKa = 3.47KK257 pKa = 11.61FEE259 pKa = 4.53IVPEE263 pKa = 4.24TTLGYY268 pKa = 8.86CQYY271 pKa = 8.71WTVAASSEE279 pKa = 4.14NQKK282 pKa = 10.27LANEE286 pKa = 5.24FINYY290 pKa = 7.46TYY292 pKa = 10.68SAEE295 pKa = 4.24SAIKK299 pKa = 10.28LLNEE303 pKa = 4.0WGGVPALKK311 pKa = 10.37RR312 pKa = 11.84EE313 pKa = 4.31LIEE316 pKa = 4.17EE317 pKa = 4.61GVAKK321 pKa = 10.39DD322 pKa = 3.61YY323 pKa = 11.4FEE325 pKa = 6.26NPMLQEE331 pKa = 4.39FADD334 pKa = 3.56MWPDD338 pKa = 3.35HH339 pKa = 6.96EE340 pKa = 4.62EE341 pKa = 3.95LSVSDD346 pKa = 4.02EE347 pKa = 3.78QTAIMDD353 pKa = 3.95TLYY356 pKa = 11.04NEE358 pKa = 4.34LMSGEE363 pKa = 4.13
Molecular weight: 40.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1M4X6S9|A0A1M4X6S9_9CLOT Uncharacterized protein OS=Lactonifactor longoviformis DSM 17459 OX=1122155 GN=SAMN02745158_01859 PE=4 SV=1
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.51GGRR28 pKa = 11.84KK29 pKa = 8.86VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
MM1 pKa = 7.67KK2 pKa = 8.72MTFQPKK8 pKa = 8.86KK9 pKa = 8.2RR10 pKa = 11.84SRR12 pKa = 11.84AKK14 pKa = 9.27VHH16 pKa = 5.81GFRR19 pKa = 11.84ARR21 pKa = 11.84MSSKK25 pKa = 10.51GGRR28 pKa = 11.84KK29 pKa = 8.86VLAARR34 pKa = 11.84RR35 pKa = 11.84LKK37 pKa = 10.68GRR39 pKa = 11.84KK40 pKa = 8.28QLSAA44 pKa = 3.9
Molecular weight: 5.04 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1447804 |
12 |
4715 |
330.5 |
36.93 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.765 ± 0.039 | 1.556 ± 0.015 |
5.257 ± 0.031 | 7.71 ± 0.041 |
3.951 ± 0.026 | 7.674 ± 0.042 |
1.7 ± 0.017 | 7.198 ± 0.038 |
6.365 ± 0.03 | 9.243 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.137 ± 0.02 | 3.892 ± 0.022 |
3.544 ± 0.021 | 3.299 ± 0.023 |
4.651 ± 0.03 | 5.778 ± 0.027 |
5.377 ± 0.03 | 6.82 ± 0.029 |
1.004 ± 0.016 | 4.079 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
